Pre Gene Modal
BGIBMGA007500
Annotation
PREDICTED:_membrane_protein_TMS1_isoform_X4_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.861
Sequence
CDS
ATGGGTGCAGTGTTGGGCCTTTGTTCTGCAGCACAGCTGGCGTGTTGCTGTGGCAGCACTGCTTGTTCACTATGTTGTTCAGCATGTCCATCTTGCACCAATTCCACCTCCTCCCGTCTTATGTATGCACTAATGCTGGTGCTGGTAACTATTGTGTGCTGTATCACTTTAGCTCCCGGATTACATAATGAACTTCAAAAGCTACCGTTTTGTACAAATGCAACTGATAGTACAGTCACGGGACTATTACCAGGAAATTTCAAGGTAGATTGTGATGAAGCGGTTGGATATTTGGCTGTTTATAGAATTACATTTGCAACATGTCTCTTCTTTCTGCTAATGGCGTTAATTATGATTGGTGTCAAATCTTCTAAAGATCCAAGAGCTGGAATTCAAAATGGTTTTTGGGCCATCAAATATTTATTGGTTATTGGTGGCATTATCGGAGCATTCTTCATCCCAGGAGGTCAATTTGCTTCGACATGGATGGTGTTTGGAATGATTGGAGGCTTCTGCTACATTGTTATCCAACTCATTCTTATCATTGATTTTGCACATTCCTGGGCTGAAATATGGGTTTCTAAATACGAAGAGACAGAATCGAAGGGATGGTATGCAGCACTACTGATTTCAATGCTGACCTGTTTTGCTCTCACCTTGACTGGCATCGTTCTGCTTTATGTTTATTACACTAAGTCCAACGGATGCGACCTATCGAAGTTCTTCATTTCGTTCAATCTGATCCTGGTGGTGGTCGCCAGCGGCGTGTCGATCTTGCCTTCGGTTCAAGAGCACCAGCCGCGCTCCGGTCTCCTGCAGTCGTCAGTCGTGTCCCTCTACGTCATGTACCTCACCTGGTCCGCGCTCTCCAACGCCTCCAAGGAATGCAACGGACTCACCGGTACATCGGATGATTCATCCTTCGACAAGCAGCGAATAATCGGGCTGTGCATTTGGGTGTGCAGCGTTCTGTACTCAAGCATAAGGACCGCCTCCGCCTCGTCCAAGATAACAATGTCCGAACACATTCTGGCCAAAGAAGGATCTACAGAGGGTGCCGACGGGGGAGAGGCGGGGCAGGGCGGGAACGAGCCTAAGGTGTTCGACAACGAAGGAGACTCCGTCGCCTACTCGTGGAGCTTCTTCCATATAATGTTCGCGCTCGCAACGCTCTACGTCATGATGACCCTCACCAATTGGTTCAACCCGAATTCGCAGCTGGCGAACGAGAATATCGCATCAATGTGGGTCAAAATAACCTCCTCTTGGATGTGCATCGGTCTCTACATTTGGACCCTGGTCGCGCCGGCCATTTTTCCGGACAGGGAATTCAATTAA
Protein
MGAVLGLCSAAQLACCCGSTACSLCCSACPSCTNSTSSRLMYALMLVLVTIVCCITLAPGLHNELQKLPFCTNATDSTVTGLLPGNFKVDCDEAVGYLAVYRITFATCLFFLLMALIMIGVKSSKDPRAGIQNGFWAIKYLLVIGGIIGAFFIPGGQFASTWMVFGMIGGFCYIVIQLILIIDFAHSWAEIWVSKYEETESKGWYAALLISMLTCFALTLTGIVLLYVYYTKSNGCDLSKFFISFNLILVVVASGVSILPSVQEHQPRSGLLQSSVVSLYVMYLTWSALSNASKECNGLTGTSDDSSFDKQRIIGLCIWVCSVLYSSIRTASASSKITMSEHILAKEGSTEGADGGEAGQGGNEPKVFDNEGDSVAYSWSFFHIMFALATLYVMMTLTNWFNPNSQLANENIASMWVKITSSWMCIGLYIWTLVAPAIFPDREFN
Summary
Uniprot
Q1EPR6
H9JDA3
A0A212EP62
A0A3S2M3L8
I4DMX0
A0A2H1VGQ6
+ More
A0A2A4J948 A0A2A4J8R5 A0A194PKZ2 A0A1E1W065 A0A1E1WDA2 A0A194R823 A0A224XNJ2 A0A224XPW2 R4FQD9 R4WJW1 A0A1B6KNF6 A0A1B6KJ62 A0A2A3E5U1 S4PY72 A0A0P4YLB8 A0A0L7QTQ8 V5G546 V9IAE7 A0A151WFJ8 A0A0P6E686 A0A0N7ZRP4 E9GGM1 A0A0P5G256 D6WM99 A0A0P5BVM3 A0A0P4ZY60 A0A0P5AEH2 A0A1B6EFW0 A0A0P5FXH3 A0A0A9YL29 A0A0A9YGY6 E2A1E8 A0A146LIL8 A0A0N8A4R2 A0A0P5U1R4 A0A0N8CFM6 A0A0P4WJC5 A0A067RBH1 A0A1B6DBU4 A0A0P5ABE3 K7IVS6 A0A0P6GYX3 A0A232F6G5 A0A0P5KID5 A0A087ZZY0 A0A310SK24 A0A195BY20 A0A0T6BGL3 A0A0P4ZGG7 A0A0N8ASC0 A0A151J6B9 A0A154PKP0 A0A195CFR8 A0A158NC97 E2B333 A0A0P5SMA2 A0A3L8E2I9 A0A1D2N529 A0A1Y1LMQ1 A0A0P5TF40 A0A195F934 A0A182UXU3 A0A1Y1LMP8 Q7QCD1 A0A1L8D977 A0A182XAU2 A0A182HSN9 A0A026W3M2 A0A182UK35 A0A1L8D941 Q178Z6 A0A182HF32 A0A1L8D9H3 Q178Z7 A0A0K8TPS3 A0A0P5YV45 A0A336MQY6 A0A0N0BJC8 A0A182K669 A0A023FM69 A0A1W4WPN1 A0A1E1XDQ4 A0A182NRK6 A0A0P5AFV7 A0A0P4WID7 A0A1W4WPM8 A0A1Q3G4J3 A0A182PZQ6 A0A1W4X0F4 A0A1Q3G4K5 G3MM96 A0A1W7R9K5 A0A023GNA1 A0A023FZR5
A0A2A4J948 A0A2A4J8R5 A0A194PKZ2 A0A1E1W065 A0A1E1WDA2 A0A194R823 A0A224XNJ2 A0A224XPW2 R4FQD9 R4WJW1 A0A1B6KNF6 A0A1B6KJ62 A0A2A3E5U1 S4PY72 A0A0P4YLB8 A0A0L7QTQ8 V5G546 V9IAE7 A0A151WFJ8 A0A0P6E686 A0A0N7ZRP4 E9GGM1 A0A0P5G256 D6WM99 A0A0P5BVM3 A0A0P4ZY60 A0A0P5AEH2 A0A1B6EFW0 A0A0P5FXH3 A0A0A9YL29 A0A0A9YGY6 E2A1E8 A0A146LIL8 A0A0N8A4R2 A0A0P5U1R4 A0A0N8CFM6 A0A0P4WJC5 A0A067RBH1 A0A1B6DBU4 A0A0P5ABE3 K7IVS6 A0A0P6GYX3 A0A232F6G5 A0A0P5KID5 A0A087ZZY0 A0A310SK24 A0A195BY20 A0A0T6BGL3 A0A0P4ZGG7 A0A0N8ASC0 A0A151J6B9 A0A154PKP0 A0A195CFR8 A0A158NC97 E2B333 A0A0P5SMA2 A0A3L8E2I9 A0A1D2N529 A0A1Y1LMQ1 A0A0P5TF40 A0A195F934 A0A182UXU3 A0A1Y1LMP8 Q7QCD1 A0A1L8D977 A0A182XAU2 A0A182HSN9 A0A026W3M2 A0A182UK35 A0A1L8D941 Q178Z6 A0A182HF32 A0A1L8D9H3 Q178Z7 A0A0K8TPS3 A0A0P5YV45 A0A336MQY6 A0A0N0BJC8 A0A182K669 A0A023FM69 A0A1W4WPN1 A0A1E1XDQ4 A0A182NRK6 A0A0P5AFV7 A0A0P4WID7 A0A1W4WPM8 A0A1Q3G4J3 A0A182PZQ6 A0A1W4X0F4 A0A1Q3G4K5 G3MM96 A0A1W7R9K5 A0A023GNA1 A0A023FZR5
Pubmed
EMBL
AB262075
BAE95628.1
BABH01030174
BABH01030175
AGBW02013542
OWR43247.1
+ More
RSAL01000046 RVE50549.1 AK402638 BAM19260.1 ODYU01002486 SOQ40029.1 NWSH01002582 PCG67923.1 PCG67924.1 KQ459602 KPI93399.1 GDQN01010722 GDQN01000100 JAT80332.1 JAT90954.1 GDQN01009455 GDQN01006147 JAT81599.1 JAT84907.1 KQ460615 KPJ13654.1 GFTR01006274 JAW10152.1 GFTR01006387 JAW10039.1 ACPB03002116 GAHY01000449 JAA77061.1 AK417900 BAN21115.1 GEBQ01026988 JAT12989.1 GEBQ01028501 JAT11476.1 KZ288387 PBC26431.1 GAIX01003793 JAA88767.1 GDIP01227179 JAI96222.1 KQ414740 KOC62038.1 GALX01003281 JAB65185.1 JR037136 JR037137 JR037138 AEY57602.1 AEY57603.1 AEY57604.1 KQ983211 KYQ46624.1 GDIP01112655 GDIQ01067419 LRGB01000725 JAL91059.1 JAN27318.1 KZS16305.1 GDIP01219185 GDIQ01249053 GDIP01131442 JAJ04217.1 JAK02672.1 GL732543 EFX81463.1 GDIQ01247700 JAK04025.1 KQ971343 EFA03337.1 GDIP01194492 JAJ28910.1 GDIP01206988 JAJ16414.1 GDIP01205087 JAJ18315.1 GEDC01028454 GEDC01000528 JAS08844.1 JAS36770.1 GDIQ01249052 JAK02673.1 GBHO01011279 GBHO01011278 GBRD01009467 JAG32325.1 JAG32326.1 JAG56357.1 GBHO01011280 GBHO01011277 GBRD01003549 JAG32324.1 JAG32327.1 JAG62272.1 GL435766 EFN72621.1 GDHC01010626 JAQ08003.1 GDIP01182641 JAJ40761.1 GDIP01120949 JAL82765.1 GDIP01136439 JAL67275.1 GDRN01068473 JAI64188.1 KK852751 KDR17149.1 GEDC01014131 GEDC01012568 JAS23167.1 JAS24730.1 GDIP01201484 JAJ21918.1 GDIQ01037306 JAN57431.1 NNAY01000843 OXU26235.1 GDIQ01184916 JAK66809.1 KQ764286 OAD54556.1 KQ976396 KYM92836.1 LJIG01000458 KRT86482.1 GDIP01217430 JAJ05972.1 GDIQ01247699 GDIP01131441 JAK04026.1 JAL72273.1 KQ979863 KYN18771.1 KQ434934 KZC11868.1 KQ977873 KYM99146.1 ADTU01011771 GL445285 EFN89882.1 GDIP01137992 JAL65722.1 QOIP01000001 RLU26673.1 LJIJ01000214 ODN00331.1 GEZM01051461 JAV74913.1 GDIP01128781 JAL74933.1 KQ981727 KYN37115.1 GEZM01051462 JAV74909.1 AAAB01008859 EAA08075.5 GFDF01011076 JAV03008.1 APCN01000311 KK107468 EZA50206.1 GFDF01011078 JAV03006.1 CH477355 EAT42822.1 JXUM01133374 JXUM01133375 JXUM01133376 KQ567972 KXJ69211.1 GFDF01011077 JAV03007.1 EAT42823.1 GDAI01001475 JAI16128.1 GDIP01052971 JAM50744.1 UFQT01002172 SSX32842.1 KQ435719 KOX78938.1 GBBK01001710 JAC22772.1 GFAC01001828 JAT97360.1 GDIP01200235 JAJ23167.1 GDIP01254828 JAI68573.1 GFDL01000324 JAV34721.1 AXCN02000002 GFDL01000321 JAV34724.1 JO842997 AEO34614.1 GFAH01000559 JAV47830.1 GBBM01000097 JAC35321.1 GBBL01001140 JAC26180.1
RSAL01000046 RVE50549.1 AK402638 BAM19260.1 ODYU01002486 SOQ40029.1 NWSH01002582 PCG67923.1 PCG67924.1 KQ459602 KPI93399.1 GDQN01010722 GDQN01000100 JAT80332.1 JAT90954.1 GDQN01009455 GDQN01006147 JAT81599.1 JAT84907.1 KQ460615 KPJ13654.1 GFTR01006274 JAW10152.1 GFTR01006387 JAW10039.1 ACPB03002116 GAHY01000449 JAA77061.1 AK417900 BAN21115.1 GEBQ01026988 JAT12989.1 GEBQ01028501 JAT11476.1 KZ288387 PBC26431.1 GAIX01003793 JAA88767.1 GDIP01227179 JAI96222.1 KQ414740 KOC62038.1 GALX01003281 JAB65185.1 JR037136 JR037137 JR037138 AEY57602.1 AEY57603.1 AEY57604.1 KQ983211 KYQ46624.1 GDIP01112655 GDIQ01067419 LRGB01000725 JAL91059.1 JAN27318.1 KZS16305.1 GDIP01219185 GDIQ01249053 GDIP01131442 JAJ04217.1 JAK02672.1 GL732543 EFX81463.1 GDIQ01247700 JAK04025.1 KQ971343 EFA03337.1 GDIP01194492 JAJ28910.1 GDIP01206988 JAJ16414.1 GDIP01205087 JAJ18315.1 GEDC01028454 GEDC01000528 JAS08844.1 JAS36770.1 GDIQ01249052 JAK02673.1 GBHO01011279 GBHO01011278 GBRD01009467 JAG32325.1 JAG32326.1 JAG56357.1 GBHO01011280 GBHO01011277 GBRD01003549 JAG32324.1 JAG32327.1 JAG62272.1 GL435766 EFN72621.1 GDHC01010626 JAQ08003.1 GDIP01182641 JAJ40761.1 GDIP01120949 JAL82765.1 GDIP01136439 JAL67275.1 GDRN01068473 JAI64188.1 KK852751 KDR17149.1 GEDC01014131 GEDC01012568 JAS23167.1 JAS24730.1 GDIP01201484 JAJ21918.1 GDIQ01037306 JAN57431.1 NNAY01000843 OXU26235.1 GDIQ01184916 JAK66809.1 KQ764286 OAD54556.1 KQ976396 KYM92836.1 LJIG01000458 KRT86482.1 GDIP01217430 JAJ05972.1 GDIQ01247699 GDIP01131441 JAK04026.1 JAL72273.1 KQ979863 KYN18771.1 KQ434934 KZC11868.1 KQ977873 KYM99146.1 ADTU01011771 GL445285 EFN89882.1 GDIP01137992 JAL65722.1 QOIP01000001 RLU26673.1 LJIJ01000214 ODN00331.1 GEZM01051461 JAV74913.1 GDIP01128781 JAL74933.1 KQ981727 KYN37115.1 GEZM01051462 JAV74909.1 AAAB01008859 EAA08075.5 GFDF01011076 JAV03008.1 APCN01000311 KK107468 EZA50206.1 GFDF01011078 JAV03006.1 CH477355 EAT42822.1 JXUM01133374 JXUM01133375 JXUM01133376 KQ567972 KXJ69211.1 GFDF01011077 JAV03007.1 EAT42823.1 GDAI01001475 JAI16128.1 GDIP01052971 JAM50744.1 UFQT01002172 SSX32842.1 KQ435719 KOX78938.1 GBBK01001710 JAC22772.1 GFAC01001828 JAT97360.1 GDIP01200235 JAJ23167.1 GDIP01254828 JAI68573.1 GFDL01000324 JAV34721.1 AXCN02000002 GFDL01000321 JAV34724.1 JO842997 AEO34614.1 GFAH01000559 JAV47830.1 GBBM01000097 JAC35321.1 GBBL01001140 JAC26180.1
Proteomes
UP000005204
UP000007151
UP000283053
UP000218220
UP000053268
UP000053240
+ More
UP000015103 UP000242457 UP000053825 UP000075809 UP000076858 UP000000305 UP000007266 UP000000311 UP000027135 UP000002358 UP000215335 UP000005203 UP000078540 UP000078492 UP000076502 UP000078542 UP000005205 UP000008237 UP000279307 UP000094527 UP000078541 UP000075903 UP000007062 UP000076407 UP000075840 UP000053097 UP000075902 UP000008820 UP000069940 UP000249989 UP000053105 UP000075881 UP000192223 UP000075884 UP000075886
UP000015103 UP000242457 UP000053825 UP000075809 UP000076858 UP000000305 UP000007266 UP000000311 UP000027135 UP000002358 UP000215335 UP000005203 UP000078540 UP000078492 UP000076502 UP000078542 UP000005205 UP000008237 UP000279307 UP000094527 UP000078541 UP000075903 UP000007062 UP000076407 UP000075840 UP000053097 UP000075902 UP000008820 UP000069940 UP000249989 UP000053105 UP000075881 UP000192223 UP000075884 UP000075886
Pfam
PF03348 Serinc
ProteinModelPortal
Q1EPR6
H9JDA3
A0A212EP62
A0A3S2M3L8
I4DMX0
A0A2H1VGQ6
+ More
A0A2A4J948 A0A2A4J8R5 A0A194PKZ2 A0A1E1W065 A0A1E1WDA2 A0A194R823 A0A224XNJ2 A0A224XPW2 R4FQD9 R4WJW1 A0A1B6KNF6 A0A1B6KJ62 A0A2A3E5U1 S4PY72 A0A0P4YLB8 A0A0L7QTQ8 V5G546 V9IAE7 A0A151WFJ8 A0A0P6E686 A0A0N7ZRP4 E9GGM1 A0A0P5G256 D6WM99 A0A0P5BVM3 A0A0P4ZY60 A0A0P5AEH2 A0A1B6EFW0 A0A0P5FXH3 A0A0A9YL29 A0A0A9YGY6 E2A1E8 A0A146LIL8 A0A0N8A4R2 A0A0P5U1R4 A0A0N8CFM6 A0A0P4WJC5 A0A067RBH1 A0A1B6DBU4 A0A0P5ABE3 K7IVS6 A0A0P6GYX3 A0A232F6G5 A0A0P5KID5 A0A087ZZY0 A0A310SK24 A0A195BY20 A0A0T6BGL3 A0A0P4ZGG7 A0A0N8ASC0 A0A151J6B9 A0A154PKP0 A0A195CFR8 A0A158NC97 E2B333 A0A0P5SMA2 A0A3L8E2I9 A0A1D2N529 A0A1Y1LMQ1 A0A0P5TF40 A0A195F934 A0A182UXU3 A0A1Y1LMP8 Q7QCD1 A0A1L8D977 A0A182XAU2 A0A182HSN9 A0A026W3M2 A0A182UK35 A0A1L8D941 Q178Z6 A0A182HF32 A0A1L8D9H3 Q178Z7 A0A0K8TPS3 A0A0P5YV45 A0A336MQY6 A0A0N0BJC8 A0A182K669 A0A023FM69 A0A1W4WPN1 A0A1E1XDQ4 A0A182NRK6 A0A0P5AFV7 A0A0P4WID7 A0A1W4WPM8 A0A1Q3G4J3 A0A182PZQ6 A0A1W4X0F4 A0A1Q3G4K5 G3MM96 A0A1W7R9K5 A0A023GNA1 A0A023FZR5
A0A2A4J948 A0A2A4J8R5 A0A194PKZ2 A0A1E1W065 A0A1E1WDA2 A0A194R823 A0A224XNJ2 A0A224XPW2 R4FQD9 R4WJW1 A0A1B6KNF6 A0A1B6KJ62 A0A2A3E5U1 S4PY72 A0A0P4YLB8 A0A0L7QTQ8 V5G546 V9IAE7 A0A151WFJ8 A0A0P6E686 A0A0N7ZRP4 E9GGM1 A0A0P5G256 D6WM99 A0A0P5BVM3 A0A0P4ZY60 A0A0P5AEH2 A0A1B6EFW0 A0A0P5FXH3 A0A0A9YL29 A0A0A9YGY6 E2A1E8 A0A146LIL8 A0A0N8A4R2 A0A0P5U1R4 A0A0N8CFM6 A0A0P4WJC5 A0A067RBH1 A0A1B6DBU4 A0A0P5ABE3 K7IVS6 A0A0P6GYX3 A0A232F6G5 A0A0P5KID5 A0A087ZZY0 A0A310SK24 A0A195BY20 A0A0T6BGL3 A0A0P4ZGG7 A0A0N8ASC0 A0A151J6B9 A0A154PKP0 A0A195CFR8 A0A158NC97 E2B333 A0A0P5SMA2 A0A3L8E2I9 A0A1D2N529 A0A1Y1LMQ1 A0A0P5TF40 A0A195F934 A0A182UXU3 A0A1Y1LMP8 Q7QCD1 A0A1L8D977 A0A182XAU2 A0A182HSN9 A0A026W3M2 A0A182UK35 A0A1L8D941 Q178Z6 A0A182HF32 A0A1L8D9H3 Q178Z7 A0A0K8TPS3 A0A0P5YV45 A0A336MQY6 A0A0N0BJC8 A0A182K669 A0A023FM69 A0A1W4WPN1 A0A1E1XDQ4 A0A182NRK6 A0A0P5AFV7 A0A0P4WID7 A0A1W4WPM8 A0A1Q3G4J3 A0A182PZQ6 A0A1W4X0F4 A0A1Q3G4K5 G3MM96 A0A1W7R9K5 A0A023GNA1 A0A023FZR5
Ontologies
GO
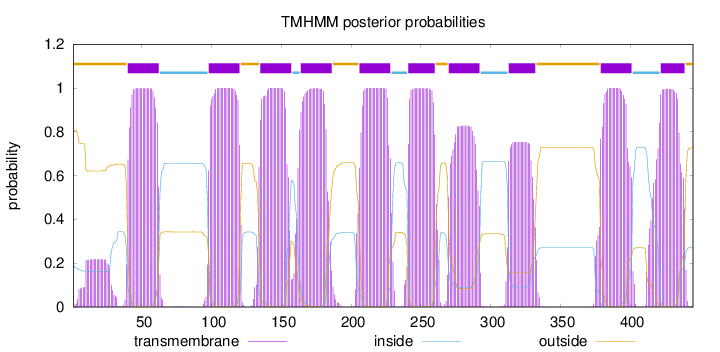
Topology
Length:
445
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
213.58289
Exp number, first 60 AAs:
25.09585
Total prob of N-in:
0.19401
POSSIBLE N-term signal
sequence
outside
1 - 39
TMhelix
40 - 62
inside
63 - 97
TMhelix
98 - 120
outside
121 - 134
TMhelix
135 - 157
inside
158 - 163
TMhelix
164 - 186
outside
187 - 205
TMhelix
206 - 228
inside
229 - 240
TMhelix
241 - 260
outside
261 - 269
TMhelix
270 - 292
inside
293 - 312
TMhelix
313 - 332
outside
333 - 378
TMhelix
379 - 401
inside
402 - 421
TMhelix
422 - 439
outside
440 - 445
Population Genetic Test Statistics
Pi
29.847911
Theta
18.76681
Tajima's D
1.049858
CLR
0.626315
CSRT
0.684115794210289
Interpretation
Uncertain
