Gene
KWMTBOMO09050 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007497
Annotation
Methylcrotonoyl-CoA_carboxylase_subunit_alpha?_mitochondrial_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 2.171
Sequence
CDS
ATGCATTATTTACGCTATCTTTATAGAAATAGTCCACTGACAACACTTCAGAACAATATAAGGTACAACCATGCACAAATCAAAGAGAAAGTTCAAAGAACACAGATCAGCAAAGTATTGATAGCAAACAGAGGGGAGATAGCATGTCGGGTCATGAGAACTGCAAAGAAATTGGGAGTCAGGACAGTGGCTGTTTATTCTGATGCCGACAGACATGCTATGCATGTTGAAATGGCAGATGAGGCTTACCACATTGGGCCTGCACCTTCCACACAAAGCTATCTAAATGCATCTAAAATTCTTGAAGTAGCTAAAAAGTCTAACAGTCAAGCTATACATCCAGGCTACGGATTTCTATCTGAAAATGTTGAATTCTGTGAAAAATGTGCTTCTGAAGATATTATATTCATTGGCCCACCACCTTCAGCTATTAGAGATATGGGTATCAAAAGTACGTCAAAAGCTATTATGTCAGCAGCCGGTGTACCCATTGTTAGAGGATACCATGGTGAAGATCAGAACATTGAGAGACTGAAAGCTGAAGCCCAAAGAATTGGTTTCCCGCTTATGATCAAGGCTGTCCGGGGGGGCGGAGGAAAGGGTATGAGAATCGCAATGACCGAATCTGATTTCCTTCCGCAATTGGAATCGGCGAAGAGAGAATCGCTTAAATCATTTGGAGATGATAATATGCTGCTCGAACAATACATTACGGATCCAAGGCATGTGGAAGTTCAGGTGTTCGCAGATATGCACGGAAATGCTGTTCATCTTTTTGAAAGAGATTGTTCAGTACAGAGACGACACCAGAAGATCATAGAAGAAGCTCCGGCACCTGGGCTTTCTGAAGAGACTCGTAGAGCGTTAGGTGAAGCCGCAGTTCGCGCCGCCAAGGCCGTGGGCTACGTAGGAGCCGGCACAGTAGAGTTCATCCTTCATAGAGTCACACACGAATTCCACTTCATGGAGATGAACACGCGTCTTCAAGTCGAACATCCAATCACGGAAATGATAACTGGTACTGATTTAGTGGAATGGCAGCTCCGCGTGGCAGCCGGTGAGCCGCTGCCCGTCACACAGGAAGACATCGTCCGGAATGGCCACGCGGTCGAGTGTAGGATATACGCAGAGGAGCCCGCGGCCGGCTTCCTGCCCAGAGCCGGTACACTGCACAGGCTAACGCAGCCTATCGCTGAAGAGTTTGTACGGGTGGAAACCGGAGTACGCGAAGGACAAGAAGTATCAGTGCACTATGATCCGATGATAGCGAAGCTGGTTGTGTGGGGACGCGATCGAACCGAGGCACTAGCGAAAACAAAGGCAAAACTTACCGAATATCAGGTAGCTGGTCTAGAAACAAACGTCAACTTCCTCCTGCGGCTGAGCGGCGCGAGCGCGTTCGTAGCGGGAGATGTTCACACAGCGTTCATACCGCAACACGAGGCCGAATTATTCCCTAAATCCGATACTGTGCTGAATGAGGACCGCGCTATTCAAGCTGCTCTGGGACACCTTCTGGTGTCACAAGCTCAACTTGCCACCAATGATAGCTGGAAAGGCATTTCTGTACCGTCTACCGCTTTCAGACCGAACTACCAGCTACAGAAGACCGTGCCATTGAGATTTGATGAAAAAGATTTCACAGTCACTGTGCAGTACACGGACGTGCCTAATACGTATCGAGTACAAGTCGGCGACGGCGAGTGGAGGAGCGTGGAAGCTTACCTCAGCAGTAATGACAAGGGCCTACGATTGAACGCTTTGATCGATGACAAGATCAGCAACATTGGATTGCTGACGTACGAACAACAAGTTCACGTGTACGACGAGACAGGGCAAACGGTTTTCGTGATACCTCATCCGAAGTTCCAAGCGGTCAGCGGATCGGACCCAGCTGCGGCTGCGAACAGCGCGTCCTCTCCCACACCCGGAGTACTTGAACGTATACTAGTTAATAAGGGGGATAAGGTGGTGAAGGGCCAGCCTCTGTTCGTGGTGATAGCAATGAAAATGGAGTATGTAGTACGTTCGCCCCGAGATGGCGTGGTGGAAGCCGTGGCACCCTTCAAACAGGGTGATGCCGTTGGCAAGGGAACCCAAGTTGTTATGCTAGAAGACCAAAGCTAA
Protein
MHYLRYLYRNSPLTTLQNNIRYNHAQIKEKVQRTQISKVLIANRGEIACRVMRTAKKLGVRTVAVYSDADRHAMHVEMADEAYHIGPAPSTQSYLNASKILEVAKKSNSQAIHPGYGFLSENVEFCEKCASEDIIFIGPPPSAIRDMGIKSTSKAIMSAAGVPIVRGYHGEDQNIERLKAEAQRIGFPLMIKAVRGGGGKGMRIAMTESDFLPQLESAKRESLKSFGDDNMLLEQYITDPRHVEVQVFADMHGNAVHLFERDCSVQRRHQKIIEEAPAPGLSEETRRALGEAAVRAAKAVGYVGAGTVEFILHRVTHEFHFMEMNTRLQVEHPITEMITGTDLVEWQLRVAAGEPLPVTQEDIVRNGHAVECRIYAEEPAAGFLPRAGTLHRLTQPIAEEFVRVETGVREGQEVSVHYDPMIAKLVVWGRDRTEALAKTKAKLTEYQVAGLETNVNFLLRLSGASAFVAGDVHTAFIPQHEAELFPKSDTVLNEDRAIQAALGHLLVSQAQLATNDSWKGISVPSTAFRPNYQLQKTVPLRFDEKDFTVTVQYTDVPNTYRVQVGDGEWRSVEAYLSSNDKGLRLNALIDDKISNIGLLTYEQQVHVYDETGQTVFVIPHPKFQAVSGSDPAAAANSASSPTPGVLERILVNKGDKVVKGQPLFVVIAMKMEYVVRSPRDGVVEAVAPFKQGDAVGKGTQVVMLEDQS
Summary
Cofactor
biotin
Similarity
Belongs to the LAMP family.
Uniprot
H9JDA0
A0A2A4JPE7
A0A194PQI7
A0A194R8S4
A0A212EP30
A0A3S2LCC8
+ More
A0A182QJ41 A0A182V0E5 A0A182N8R7 A0A182TEY7 A0A2M4CNV0 A0A182I0R8 A0A1S4H3D0 Q7Q0P5 A0A182XM82 D3TLV3 A0A1J1HUW4 A0A182JTC5 W5JDL2 A0A182R6W9 A0A2M4CNS1 A0A182P883 A0A182FMW2 A0A1A9WRM8 A0A2M4BFS9 A0A2M4AAT8 A0A1I8N9P6 A0A182YHD7 A0A3B5QHG4 T1PFQ2 A0A0K8USN4 A0A2M3Z4U3 A0A2M3Z509 A0A182M8D6 A0A0A1XK85 A0A034WU43 A0A182VSX4 A0A3P9QH10 A0A182IIX9 A0A096MCM5 A0A087YMP0 A0A3B3WK20 A0A3Q1JBZ6 A0A3Q0T7S2 K7JAC5 Q16TC2 A0A3Q2GJS0 A0A3Q0T208 A0A232F0P3 A0A3B3W0Z3 A0A1Q3EY21 H3BFE7 Q16TC4 E9IZL2 A0A2T7PCT5 A0A0L0BT33 I3JNV9 B4JXZ5 A0A2D0RTX9 A0A444TZ89 A0A3Q3S6Y7 H2S6P6 A0A3L8DJI6 A0A3Q3KH11 A0A3Q2WE85 A0A3P8RCQ2 A0A3P9CEM8 W8B407 A0A3B3DG69 B4PP29 H3CMT4 A0A1A8B6Y3 H3C4J4 A0A026WX95 M3ZEG5 A0A2I4C7P5 H3C477 A0A1A8VIH9 A0A3Q2XTP4 A0A1A8BL62 A0A1A8E9H4 A0A3P8RCD5 A0A1A7YBA8 W5K1X5 F6YSD6 A0A1A8IU07 F4WDJ9 Q6DIZ7 A0A1A7X4P4 G3NRZ8 D6WKZ0 A0A195FRS7 A0A1A8NYA7 N6TU04 A0A3Q1F6I0 B4NGN7 A0A1A8LGW9 A0A1W4VLW8 B4LVX1
A0A182QJ41 A0A182V0E5 A0A182N8R7 A0A182TEY7 A0A2M4CNV0 A0A182I0R8 A0A1S4H3D0 Q7Q0P5 A0A182XM82 D3TLV3 A0A1J1HUW4 A0A182JTC5 W5JDL2 A0A182R6W9 A0A2M4CNS1 A0A182P883 A0A182FMW2 A0A1A9WRM8 A0A2M4BFS9 A0A2M4AAT8 A0A1I8N9P6 A0A182YHD7 A0A3B5QHG4 T1PFQ2 A0A0K8USN4 A0A2M3Z4U3 A0A2M3Z509 A0A182M8D6 A0A0A1XK85 A0A034WU43 A0A182VSX4 A0A3P9QH10 A0A182IIX9 A0A096MCM5 A0A087YMP0 A0A3B3WK20 A0A3Q1JBZ6 A0A3Q0T7S2 K7JAC5 Q16TC2 A0A3Q2GJS0 A0A3Q0T208 A0A232F0P3 A0A3B3W0Z3 A0A1Q3EY21 H3BFE7 Q16TC4 E9IZL2 A0A2T7PCT5 A0A0L0BT33 I3JNV9 B4JXZ5 A0A2D0RTX9 A0A444TZ89 A0A3Q3S6Y7 H2S6P6 A0A3L8DJI6 A0A3Q3KH11 A0A3Q2WE85 A0A3P8RCQ2 A0A3P9CEM8 W8B407 A0A3B3DG69 B4PP29 H3CMT4 A0A1A8B6Y3 H3C4J4 A0A026WX95 M3ZEG5 A0A2I4C7P5 H3C477 A0A1A8VIH9 A0A3Q2XTP4 A0A1A8BL62 A0A1A8E9H4 A0A3P8RCD5 A0A1A7YBA8 W5K1X5 F6YSD6 A0A1A8IU07 F4WDJ9 Q6DIZ7 A0A1A7X4P4 G3NRZ8 D6WKZ0 A0A195FRS7 A0A1A8NYA7 N6TU04 A0A3Q1F6I0 B4NGN7 A0A1A8LGW9 A0A1W4VLW8 B4LVX1
Pubmed
19121390
26354079
22118469
12364791
20353571
20920257
+ More
23761445 25315136 25244985 23542700 25830018 25348373 20075255 17510324 28648823 9215903 21282665 26108605 25186727 17994087 21551351 30249741 24495485 29451363 17550304 15496914 24508170 25329095 20431018 21719571 18362917 19820115 23537049
23761445 25315136 25244985 23542700 25830018 25348373 20075255 17510324 28648823 9215903 21282665 26108605 25186727 17994087 21551351 30249741 24495485 29451363 17550304 15496914 24508170 25329095 20431018 21719571 18362917 19820115 23537049
EMBL
BABH01030178
BABH01030179
BABH01030180
BABH01030181
NWSH01000837
PCG73927.1
+ More
KQ459602 KPI93395.1 KQ460615 KPJ13650.1 AGBW02013542 OWR43252.1 RSAL01000046 RVE50551.1 AXCN02001862 GGFL01002819 MBW66997.1 APCN01005251 AAAB01008980 EAA14349.4 EZ422405 ADD18681.1 CVRI01000020 CRK91156.1 ADMH02001476 ETN62447.1 GGFL01002798 MBW66976.1 GGFJ01002769 MBW51910.1 GGFK01004549 MBW37870.1 KA647581 AFP62210.1 GDHF01022723 JAI29591.1 GGFM01002788 MBW23539.1 GGFM01002787 MBW23538.1 AXCM01000430 GBXI01003329 JAD10963.1 GAKP01000798 JAC58154.1 AYCK01017815 AAZX01000433 CH477653 EAT37748.1 NNAY01001365 OXU24234.1 GFDL01014843 JAV20202.1 AFYH01019143 AFYH01019144 AFYH01019145 AFYH01019146 EAT37746.1 GL767232 EFZ14002.1 PZQS01000004 PVD31231.1 JRES01001410 KNC23148.1 AERX01054195 CH916377 EDV90557.1 SCEB01215691 RXM28264.1 QOIP01000007 RLU20496.1 GAMC01010675 JAB95880.1 CM000160 EDW99265.1 HADY01023951 SBP62436.1 KK107072 EZA60675.1 HADY01000604 HAEJ01018915 SBS59372.1 HADZ01003682 SBP67623.1 HAEA01014113 SBQ42593.1 HADX01005284 SBP27516.1 AAMC01063454 HAED01014381 HAEE01000094 SBR00826.1 GL888087 EGI67779.1 BC075388 AAH75388.1 HADW01011528 SBP12928.1 KQ971343 EFA03540.1 KQ981285 KYN43158.1 HAEI01000889 HAEH01009496 SBR73747.1 APGK01018863 KB740085 ENN81513.1 CH964272 EDW84384.1 HAEF01006728 SBR44110.1 CH940650 EDW67576.1
KQ459602 KPI93395.1 KQ460615 KPJ13650.1 AGBW02013542 OWR43252.1 RSAL01000046 RVE50551.1 AXCN02001862 GGFL01002819 MBW66997.1 APCN01005251 AAAB01008980 EAA14349.4 EZ422405 ADD18681.1 CVRI01000020 CRK91156.1 ADMH02001476 ETN62447.1 GGFL01002798 MBW66976.1 GGFJ01002769 MBW51910.1 GGFK01004549 MBW37870.1 KA647581 AFP62210.1 GDHF01022723 JAI29591.1 GGFM01002788 MBW23539.1 GGFM01002787 MBW23538.1 AXCM01000430 GBXI01003329 JAD10963.1 GAKP01000798 JAC58154.1 AYCK01017815 AAZX01000433 CH477653 EAT37748.1 NNAY01001365 OXU24234.1 GFDL01014843 JAV20202.1 AFYH01019143 AFYH01019144 AFYH01019145 AFYH01019146 EAT37746.1 GL767232 EFZ14002.1 PZQS01000004 PVD31231.1 JRES01001410 KNC23148.1 AERX01054195 CH916377 EDV90557.1 SCEB01215691 RXM28264.1 QOIP01000007 RLU20496.1 GAMC01010675 JAB95880.1 CM000160 EDW99265.1 HADY01023951 SBP62436.1 KK107072 EZA60675.1 HADY01000604 HAEJ01018915 SBS59372.1 HADZ01003682 SBP67623.1 HAEA01014113 SBQ42593.1 HADX01005284 SBP27516.1 AAMC01063454 HAED01014381 HAEE01000094 SBR00826.1 GL888087 EGI67779.1 BC075388 AAH75388.1 HADW01011528 SBP12928.1 KQ971343 EFA03540.1 KQ981285 KYN43158.1 HAEI01000889 HAEH01009496 SBR73747.1 APGK01018863 KB740085 ENN81513.1 CH964272 EDW84384.1 HAEF01006728 SBR44110.1 CH940650 EDW67576.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000075886 UP000075903 UP000075884 UP000075902 UP000075840 UP000007062 UP000076407 UP000183832 UP000075881 UP000000673 UP000075900 UP000075885 UP000069272 UP000091820 UP000095301 UP000076408 UP000002852 UP000075883 UP000075920 UP000242638 UP000075880 UP000028760 UP000261480 UP000265040 UP000261340 UP000002358 UP000008820 UP000265020 UP000215335 UP000261500 UP000008672 UP000245119 UP000037069 UP000005207 UP000001070 UP000221080 UP000261640 UP000005226 UP000279307 UP000261600 UP000264840 UP000265100 UP000265160 UP000261560 UP000002282 UP000007303 UP000053097 UP000192220 UP000264820 UP000018467 UP000008143 UP000007755 UP000007635 UP000007266 UP000078541 UP000019118 UP000257200 UP000007798 UP000192221 UP000008792
UP000075886 UP000075903 UP000075884 UP000075902 UP000075840 UP000007062 UP000076407 UP000183832 UP000075881 UP000000673 UP000075900 UP000075885 UP000069272 UP000091820 UP000095301 UP000076408 UP000002852 UP000075883 UP000075920 UP000242638 UP000075880 UP000028760 UP000261480 UP000265040 UP000261340 UP000002358 UP000008820 UP000265020 UP000215335 UP000261500 UP000008672 UP000245119 UP000037069 UP000005207 UP000001070 UP000221080 UP000261640 UP000005226 UP000279307 UP000261600 UP000264840 UP000265100 UP000265160 UP000261560 UP000002282 UP000007303 UP000053097 UP000192220 UP000264820 UP000018467 UP000008143 UP000007755 UP000007635 UP000007266 UP000078541 UP000019118 UP000257200 UP000007798 UP000192221 UP000008792
PRIDE
Pfam
Interpro
IPR011761
ATP-grasp
+ More
IPR011054 Rudment_hybrid_motif
IPR005482 Biotin_COase_C
IPR001882 Biotin_BS
IPR016185 PreATP-grasp_dom_sf
IPR005479 CbamoylP_synth_lsu-like_ATP-bd
IPR000089 Biotin_lipoyl
IPR011053 Single_hybrid_motif
IPR011764 Biotin_carboxylation_dom
IPR005481 BC-like_N
IPR013815 ATP_grasp_subdomain_1
IPR003010 C-N_Hydrolase
IPR036526 C-N_Hydrolase_sf
IPR002000 Lysosome-assoc_membr_glycop
IPR019176 Cytochrome_B561-rel
IPR040122 Importin_beta
IPR011989 ARM-like
IPR001494 Importin-beta_N
IPR016024 ARM-type_fold
IPR011054 Rudment_hybrid_motif
IPR005482 Biotin_COase_C
IPR001882 Biotin_BS
IPR016185 PreATP-grasp_dom_sf
IPR005479 CbamoylP_synth_lsu-like_ATP-bd
IPR000089 Biotin_lipoyl
IPR011053 Single_hybrid_motif
IPR011764 Biotin_carboxylation_dom
IPR005481 BC-like_N
IPR013815 ATP_grasp_subdomain_1
IPR003010 C-N_Hydrolase
IPR036526 C-N_Hydrolase_sf
IPR002000 Lysosome-assoc_membr_glycop
IPR019176 Cytochrome_B561-rel
IPR040122 Importin_beta
IPR011989 ARM-like
IPR001494 Importin-beta_N
IPR016024 ARM-type_fold
SUPFAM
Gene 3D
ProteinModelPortal
H9JDA0
A0A2A4JPE7
A0A194PQI7
A0A194R8S4
A0A212EP30
A0A3S2LCC8
+ More
A0A182QJ41 A0A182V0E5 A0A182N8R7 A0A182TEY7 A0A2M4CNV0 A0A182I0R8 A0A1S4H3D0 Q7Q0P5 A0A182XM82 D3TLV3 A0A1J1HUW4 A0A182JTC5 W5JDL2 A0A182R6W9 A0A2M4CNS1 A0A182P883 A0A182FMW2 A0A1A9WRM8 A0A2M4BFS9 A0A2M4AAT8 A0A1I8N9P6 A0A182YHD7 A0A3B5QHG4 T1PFQ2 A0A0K8USN4 A0A2M3Z4U3 A0A2M3Z509 A0A182M8D6 A0A0A1XK85 A0A034WU43 A0A182VSX4 A0A3P9QH10 A0A182IIX9 A0A096MCM5 A0A087YMP0 A0A3B3WK20 A0A3Q1JBZ6 A0A3Q0T7S2 K7JAC5 Q16TC2 A0A3Q2GJS0 A0A3Q0T208 A0A232F0P3 A0A3B3W0Z3 A0A1Q3EY21 H3BFE7 Q16TC4 E9IZL2 A0A2T7PCT5 A0A0L0BT33 I3JNV9 B4JXZ5 A0A2D0RTX9 A0A444TZ89 A0A3Q3S6Y7 H2S6P6 A0A3L8DJI6 A0A3Q3KH11 A0A3Q2WE85 A0A3P8RCQ2 A0A3P9CEM8 W8B407 A0A3B3DG69 B4PP29 H3CMT4 A0A1A8B6Y3 H3C4J4 A0A026WX95 M3ZEG5 A0A2I4C7P5 H3C477 A0A1A8VIH9 A0A3Q2XTP4 A0A1A8BL62 A0A1A8E9H4 A0A3P8RCD5 A0A1A7YBA8 W5K1X5 F6YSD6 A0A1A8IU07 F4WDJ9 Q6DIZ7 A0A1A7X4P4 G3NRZ8 D6WKZ0 A0A195FRS7 A0A1A8NYA7 N6TU04 A0A3Q1F6I0 B4NGN7 A0A1A8LGW9 A0A1W4VLW8 B4LVX1
A0A182QJ41 A0A182V0E5 A0A182N8R7 A0A182TEY7 A0A2M4CNV0 A0A182I0R8 A0A1S4H3D0 Q7Q0P5 A0A182XM82 D3TLV3 A0A1J1HUW4 A0A182JTC5 W5JDL2 A0A182R6W9 A0A2M4CNS1 A0A182P883 A0A182FMW2 A0A1A9WRM8 A0A2M4BFS9 A0A2M4AAT8 A0A1I8N9P6 A0A182YHD7 A0A3B5QHG4 T1PFQ2 A0A0K8USN4 A0A2M3Z4U3 A0A2M3Z509 A0A182M8D6 A0A0A1XK85 A0A034WU43 A0A182VSX4 A0A3P9QH10 A0A182IIX9 A0A096MCM5 A0A087YMP0 A0A3B3WK20 A0A3Q1JBZ6 A0A3Q0T7S2 K7JAC5 Q16TC2 A0A3Q2GJS0 A0A3Q0T208 A0A232F0P3 A0A3B3W0Z3 A0A1Q3EY21 H3BFE7 Q16TC4 E9IZL2 A0A2T7PCT5 A0A0L0BT33 I3JNV9 B4JXZ5 A0A2D0RTX9 A0A444TZ89 A0A3Q3S6Y7 H2S6P6 A0A3L8DJI6 A0A3Q3KH11 A0A3Q2WE85 A0A3P8RCQ2 A0A3P9CEM8 W8B407 A0A3B3DG69 B4PP29 H3CMT4 A0A1A8B6Y3 H3C4J4 A0A026WX95 M3ZEG5 A0A2I4C7P5 H3C477 A0A1A8VIH9 A0A3Q2XTP4 A0A1A8BL62 A0A1A8E9H4 A0A3P8RCD5 A0A1A7YBA8 W5K1X5 F6YSD6 A0A1A8IU07 F4WDJ9 Q6DIZ7 A0A1A7X4P4 G3NRZ8 D6WKZ0 A0A195FRS7 A0A1A8NYA7 N6TU04 A0A3Q1F6I0 B4NGN7 A0A1A8LGW9 A0A1W4VLW8 B4LVX1
PDB
3U9T
E-value=1.63296e-142,
Score=1300
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Lysosome membrane
Nucleus
Nucleus
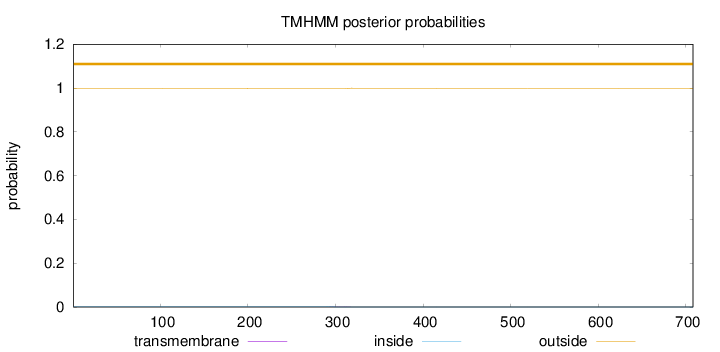
Length:
708
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02823
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00184
outside
1 - 708
Population Genetic Test Statistics
Pi
215.681411
Theta
174.846305
Tajima's D
0.579442
CLR
0.768053
CSRT
0.543422828858557
Interpretation
Possibly Positive selection
