Gene
KWMTBOMO09049
Pre Gene Modal
BGIBMGA007587
Annotation
PREDICTED:_flap_endonuclease_GEN_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.812
Sequence
CDS
ATGGGTATAAAAGGTTTGTGGTCTGTGCTGACTCCTTTCAGTGAAAAGAAATCCTTGCATGAATTACGGGGGGAAACAATTGCAGTAGACCTCTCTGGTTGGGTCTGTGATAGTCAGAATGTTACGGAACACCATGTCCAACCGAAATTATATCTTAGAAACCTGTTCTTCAGAACGGTGTATCTGCTATTGGCTGAAATAAATCCTATATTTGTATTGGAAGGAGATGCACCAGAACTCAAAAGAGATGTAATGGCTACTCGAAATGCTGTCCAATTTCGTGGTGCCGCACCCAGATCTGAGAAGGCTTGTTCAAGTGAAAAACTGCCTAATGTTTCCAGAAAGAGATTTAAAAATGTTTTAAAGGAATGTGAAACTCTTCTCAAGAGTATGGGAGTGATATGTCTTAAAGGTAGCGGAGAAGCTGAAGCAACATGTGCCCAATTAAATGCTGAAGGTTTCGTCGATGCAGTTGTCTCCCAAGATTCGGATTGTTTTGCTTACGGAGCGAAACGAGTGTACCGAAACTTTAGCGTGTCGAGTTCAGCCGGCGGAGGTGCGATGCAGGGCTCTGTTGACTGCTATGACGCCGAAAAACTGTACAAGTCGAATGGGTTCGGCCGCAACAAAATGGTGGCGTTGGCTTTGCTTTGCGGGTGTGACTACGTCGTGGGCGCGTGTGGTTCGTCCATAACGACGGCCGTGCAATTTTTGCACACTGTTGCTGATGATGATGTAATTCCTAGGTTATTATCGTGGGTGTCGGATCCCGATTGTTACGAGAGGCGGGCTCGCTGGGCGTCCGCCCCGGGACGCTGCGACCGCTGCGGTCACGTCGGACGCACACACCTCAGGAATGGATGCCCCGTTTGCGCCACGGACCGAGGCTGCAACGATCTCGGACATAAGTCGAAAGTAGCGGAAGCGAAGCGAGAGCTGTCACTTCGCAGCCGGGCCCTGTCGAGCGGCGTGCTCTTCCCCGAGCCGAGAGTTATGAAGGAGTTTTTGAAGCCCACAATTGAAAAAATTGAGTTGGACAGCTTGAAAACGCCGGTTCCGAGTTTGATACAGTTTGTGAAATTAATGGTGAAGAAACTTGATTGGTCGGAACGGTACTGCGTCGAGAAATTCTTGCCGTTGCTAACGAAATTTCATTTACAAGAACGCGTTCCGTCTAGAACCGTGCAACCCATTCGTATAAAGAAAAAGCGGAACCCGAGAGGTGTTCCGAGTTACGAAGTGGTGTGGGCTGACGTTAACGGAGTATACGAAGCACTAATTCCTGATGATCAATTTGAAGAAGATGAAGATCCCTCTGTTGTTTGGACGTCAACAGAGAGACAAGATCTCATGCGGCAGTTTTATCCGAAGATAGTTGAGGCATTCGAAGAATCTATAAAGAAACCTCTAAAAGCAAAGAAATCAAGAACCAAGAAAAAGAAAGAAGAAAAAGAAAATATACAGGAAGATAAACCGAAAAGGCGTTATAATAGAAAACCTAAAAATAACGTTGAAATTTTAATATTAAATGAATCTAATAAACATTCGAAATTTGAATCGAATTCGAGCAAGCTGAATGTCAGTGTTTCCGTGAATAAATTAAAAAGGAAGTTGAAAAACACGAAGAAGTCCACACAAAAAACCATGGACAGTTTTATTGCGAAGAAAAAGAAGAAATCGTTGCGTAACAGTTTGGTCTTGAGTGTGAGGAGGAGCTTTAAGAATTTGTCCATAGCTGAGGATAAAAAAATAAATAAAAATAAAAACTTTTTGAGCATACTGAATAAATCAGACGAACTAGATCACAGTGACTTGTCAGGTATTATAGAGACTATAATTTCGAGATCACCCGTAGTCGAAACAGCCAAAGTGGAAAACAAAACGTTCAAACTTGTATTCGATAAATATTCAACGCCGACAAAATTCGCTTTACGTAAAACTGTACTGAGCGATATACAGAATAATTGTTCGACACCGAAAGACAGTCCGAAGAACAGTTTTAATGCTTCAATGAAATCTAAATTGAATACGAGTTATTTCTTTGATAAATTGACTGAGGAAAGGGACGCCTTCGAAATGTCGCTCGATTACAAGCACAATAGCGCTGTTATTGTTGAATATGATAAGACTTTAGATTATAGTTTGCCCGAAGTCTGGTGA
Protein
MGIKGLWSVLTPFSEKKSLHELRGETIAVDLSGWVCDSQNVTEHHVQPKLYLRNLFFRTVYLLLAEINPIFVLEGDAPELKRDVMATRNAVQFRGAAPRSEKACSSEKLPNVSRKRFKNVLKECETLLKSMGVICLKGSGEAEATCAQLNAEGFVDAVVSQDSDCFAYGAKRVYRNFSVSSSAGGGAMQGSVDCYDAEKLYKSNGFGRNKMVALALLCGCDYVVGACGSSITTAVQFLHTVADDDVIPRLLSWVSDPDCYERRARWASAPGRCDRCGHVGRTHLRNGCPVCATDRGCNDLGHKSKVAEAKRELSLRSRALSSGVLFPEPRVMKEFLKPTIEKIELDSLKTPVPSLIQFVKLMVKKLDWSERYCVEKFLPLLTKFHLQERVPSRTVQPIRIKKKRNPRGVPSYEVVWADVNGVYEALIPDDQFEEDEDPSVVWTSTERQDLMRQFYPKIVEAFEESIKKPLKAKKSRTKKKKEEKENIQEDKPKRRYNRKPKNNVEILILNESNKHSKFESNSSKLNVSVSVNKLKRKLKNTKKSTQKTMDSFIAKKKKKSLRNSLVLSVRRSFKNLSIAEDKKINKNKNFLSILNKSDELDHSDLSGIIETIISRSPVVETAKVENKTFKLVFDKYSTPTKFALRKTVLSDIQNNCSTPKDSPKNSFNASMKSKLNTSYFFDKLTEERDAFEMSLDYKHNSAVIVEYDKTLDYSLPEVW
Summary
Uniprot
Proteomes
PRIDE
Interpro
ProteinModelPortal
PDB
5T9J
E-value=8.18285e-57,
Score=561
Ontologies
GO
PANTHER
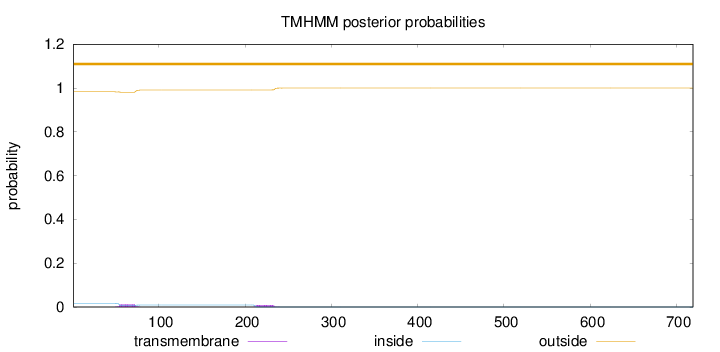
Topology
Length:
719
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.45679
Exp number, first 60 AAs:
0.08667
Total prob of N-in:
0.01740
outside
1 - 719
Population Genetic Test Statistics
Pi
199.651143
Theta
155.838134
Tajima's D
-1.461939
CLR
1010.629716
CSRT
0.0657967101644918
Interpretation
Possibly Positive selection
