Gene
KWMTBOMO09034 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007594
Annotation
PREDICTED:_putative_ATP-dependent_RNA_helicase_Pl10_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.121
Sequence
CDS
ATGAGTAATGTTACCAACCAAAATGGAACAGGTCTAGAGCAGCAGCTTGCTGGTCTGGACTTGCAGCCCCAGGCTCCTAAGAGTACTGGTCGCTACATACCCCCGCATCTGCGTAGGCAGTTGCAGGCAACCTCTGATCAAGGGGAAGAGTCTAAGCGCTCAAGCTTAGACACCCGTCCTAGTGAGTCACGGGACAACCGTTCATCATTCGGAGGCAGCTCTAGAGGCTACGACCGTGGTGGTCGGCGCGACGATCGTGATCGTGACCGCGAACGTGATTACGATCGCGGATACGAAGCGCGACACCAGCGCAGGGACCGCGGTCGAGACAGTGGATCTCAGAATGGCGAACCAGAGCGTTGGAATGAATCCGAGAAATGGAATGATCGTGAACGCGAACGGGACCGACGCGGGCCTCGCGACGTGCGACGTGAGGAGGATTATGAACCACCGGCACCACGGAATGACCGCTGGAAAGAGCCGGAGCCAAGAGCTGAAGAGCGTTCCAATTCACGCTGGCCTTCTGACGATGTGCGCCGAACTAGATCTCGCAAAGACGAGAATGATTGGACAGTACCGTTACCTCGCGATGAACGACAGGAACTATTGCTGTTCGGCACTGGCAACACCGGGATCAATTTCTCCAAATACGAGGATATACCGGTTGAGGCCAGCGGAGACCGCGTACCAGACTGCATCACCAGTTTTGAGGATGTTAACCTGACAGAACTAATGCGGACAAATATATCTCTCGCTCGCTATGACAAGCCGACGCCGGTACAGAAGTATGCGATTCCGATTATTTTGTCACGTAGAGACGTGATGGCGTGTGCACAGACGGGATCTGGAAAAACCGCAGCCTTTCTTGTTCCCATACTTAACCAAATGTATGAAGCAGGACCTGTGAAACACATGGGACCTTATAATAAACGGAAACAATATCCTTTGGGCTTGGTGTTAGCCCCAACGCGAGAGCTCGCCACCCAAATTTTTGATGAAGCTAGAAAATTTGCCTATCGATCACGAGTCCGACCCTGTGTGGTGTATGGAGGCTCACCTATTAATGATCAGTTCCGTGAATTAGAGTCAGGATGTCATCTGTTAGTAGCCACTCCTGGGCGTTTGGTAGACATGCTGGCTCGTGGCAGAGTTGCATTGGATCACTGTCGCCATCTTGTACTCGACGAAGCCGACCGCATGTTGGACATGGGTTTTGAGCCACAGATCCGTAAGATTGTAGAGTGCCACACGATGCCCAAGACTGGCGAACGTCAAACTCTTATGTTTTCTGCCACTTTCCCAAAACAGATTCAACTTTTGGCTCAGGATTTCTTATACAATTATGTGTTCCTTGCTGTTGGACGTGTAGGCTCAACGTCAGAAAATATTACACAGAAGGTTGTTTGGGTAGACGAAATGGACAAGCGTTCCTTTTTATTGGATTTACTAAATGCATCAAACTTGCTACAACGTGCACGTCCCGAAGAGGATCAACTTATACTTGTGTTTGTGGAAACTAAGAAAGGTGCGGATCAACTAGAAGAATATTTATATTCCCAAGGCTACCCGGTAACATCGATCCATGGAGACCGTAACCAACGTGAACGTGAAGATGCTTTACGACGTTTTCGAACTGGACAAACACCAATACTTGTGGCAACAGCTGTCGCTGCAAGAGGCCTAGACATACCGCACGTACGTCACGTAATAAATTTTGATCTACCATCTGACGTGGAAGAATACGTCCATCGTATCGGTCGTACAGGTCGTATGGGTAACCTCGGTGTGGCTACTTCCTTCTTTAATGACACAAACCGTGGTTTGGCACGAGACCTCGTCGAACTGCTCGTCGAGGCTAAACAAGACGTACCTAATTGGCTTACTAGCACCGCAGCTGACGGACGGCTTGGCGGCGGCGGTCGGCGCACTGGCTCTCGTTCTGGGAATGCTCGTTACGGCGGTAGCGGCTTCGGCTCAAGGGACTTCCGTACACAACCTCGTCAGGCGGCACGTTCTGCAGGAGCTGGCGGTATCGGTTCCGGCTTTGGCTATGACGTGGGCTCATATGGTGGTAACTATGGCGGGTCGTACGGCGGAGGAGGCGGAGGCGGTAACAGCGGAGGTCCCGACTGGTGGGATTCCTAA
Protein
MSNVTNQNGTGLEQQLAGLDLQPQAPKSTGRYIPPHLRRQLQATSDQGEESKRSSLDTRPSESRDNRSSFGGSSRGYDRGGRRDDRDRDRERDYDRGYEARHQRRDRGRDSGSQNGEPERWNESEKWNDRERERDRRGPRDVRREEDYEPPAPRNDRWKEPEPRAEERSNSRWPSDDVRRTRSRKDENDWTVPLPRDERQELLLFGTGNTGINFSKYEDIPVEASGDRVPDCITSFEDVNLTELMRTNISLARYDKPTPVQKYAIPIILSRRDVMACAQTGSGKTAAFLVPILNQMYEAGPVKHMGPYNKRKQYPLGLVLAPTRELATQIFDEARKFAYRSRVRPCVVYGGSPINDQFRELESGCHLLVATPGRLVDMLARGRVALDHCRHLVLDEADRMLDMGFEPQIRKIVECHTMPKTGERQTLMFSATFPKQIQLLAQDFLYNYVFLAVGRVGSTSENITQKVVWVDEMDKRSFLLDLLNASNLLQRARPEEDQLILVFVETKKGADQLEEYLYSQGYPVTSIHGDRNQREREDALRRFRTGQTPILVATAVAARGLDIPHVRHVINFDLPSDVEEYVHRIGRTGRMGNLGVATSFFNDTNRGLARDLVELLVEAKQDVPNWLTSTAADGRLGGGGRRTGSRSGNARYGGSGFGSRDFRTQPRQAARSAGAGGIGSGFGYDVGSYGGNYGGSYGGGGGGGNSGGPDWWDS
Summary
Similarity
Belongs to the DEAD box helicase family.
Uniprot
A0A2A4K7F1
A0A2A4K6T1
A0A212EUG3
A0A437BJM2
A0A194PQH6
A0A194R8R4
+ More
H9JDJ7 A0A0L7LDT1 A0A0T6B0A8 A0A2H1W3F8 A0A0C9RAD5 A0A2J7RT20 A0A067R0G3 E9HD16 A0A310SAB9 A0A1B6M6Z0 A0A0L7QZA0 A0A1B6M1Y3 A0A0P5G8S5 A0A0P5G0W9 A0A0P5FWM3 A0A0P5UIY8 A0A0N8DD80 A0A0P5DLD9 A0A0P6I4V5 A0A0P5JG94 A0A0P5FSJ2 A0A0P5YZE3 A0A0N8DVH9 A0A2A3E6Y0 A0A0P5RNR6 A0A0P6BLR4 A0A0P6CZT9 V9IJF1 A0A0B4J2N5 A0A0N7ZQW4 A0A0P5MFB6 A0A0P5TAG6 A0A0P5S0Z4 A0A0P5IHA1 A0A0N7ZJE4 A0A0N8DC50 A0A2A3E8C1 A0A0P5G8K0 A0A0P5WC41 A0A0P5HM30 A0A0P5T0E3 A0A1B6FKZ8 A0A1B6I237 A0A0P5Q1M2 A0A154P1K4 A0A195FTR0 F4WAH3 J9K0U9 A0A158NVN2 A0A224X8R8 A0A195CB22 A0A151X5N7 A0A1B6C3L3 A0A0B4J2R6 A0A0P4W3R8 T1IWF1 A0A444SNF3 A0A1A9W9C9 A0A336MFD9 A0A0R3NP78 B5DWF0 A0A3R7PL42 A0A3B0JLC3 U5EU41 B0L0X1 A0A1A9VVY0 W8B905 B4G457 A0A433TP95
H9JDJ7 A0A0L7LDT1 A0A0T6B0A8 A0A2H1W3F8 A0A0C9RAD5 A0A2J7RT20 A0A067R0G3 E9HD16 A0A310SAB9 A0A1B6M6Z0 A0A0L7QZA0 A0A1B6M1Y3 A0A0P5G8S5 A0A0P5G0W9 A0A0P5FWM3 A0A0P5UIY8 A0A0N8DD80 A0A0P5DLD9 A0A0P6I4V5 A0A0P5JG94 A0A0P5FSJ2 A0A0P5YZE3 A0A0N8DVH9 A0A2A3E6Y0 A0A0P5RNR6 A0A0P6BLR4 A0A0P6CZT9 V9IJF1 A0A0B4J2N5 A0A0N7ZQW4 A0A0P5MFB6 A0A0P5TAG6 A0A0P5S0Z4 A0A0P5IHA1 A0A0N7ZJE4 A0A0N8DC50 A0A2A3E8C1 A0A0P5G8K0 A0A0P5WC41 A0A0P5HM30 A0A0P5T0E3 A0A1B6FKZ8 A0A1B6I237 A0A0P5Q1M2 A0A154P1K4 A0A195FTR0 F4WAH3 J9K0U9 A0A158NVN2 A0A224X8R8 A0A195CB22 A0A151X5N7 A0A1B6C3L3 A0A0B4J2R6 A0A0P4W3R8 T1IWF1 A0A444SNF3 A0A1A9W9C9 A0A336MFD9 A0A0R3NP78 B5DWF0 A0A3R7PL42 A0A3B0JLC3 U5EU41 B0L0X1 A0A1A9VVY0 W8B905 B4G457 A0A433TP95
Pubmed
EMBL
NWSH01000073
PCG79946.1
PCG79947.1
AGBW02012390
OWR45135.1
RSAL01000046
+ More
RVE50559.1 KQ459602 KPI93385.1 KQ460615 KPJ13640.1 BABH01030221 JTDY01001556 KOB73544.1 LJIG01016384 KRT80762.1 ODYU01006090 SOQ47630.1 GBYB01003873 GBYB01013404 JAG73640.1 JAG83171.1 NEVH01000249 PNF43980.1 KK853131 KDR10924.1 GL732622 EFX70372.1 KQ762360 OAD55924.1 GEBQ01008313 JAT31664.1 KQ414681 KOC63923.1 GEBQ01010055 JAT29922.1 GDIQ01252053 JAJ99671.1 GDIQ01249041 JAK02684.1 GDIQ01250641 JAK01084.1 GDIP01112230 JAL91484.1 GDIP01045207 JAM58508.1 GDIP01160732 JAJ62670.1 GDIQ01026257 JAN68480.1 GDIQ01200394 JAK51331.1 GDIQ01252052 JAJ99672.1 GDIP01052899 JAM50816.1 GDIP01158430 GDIQ01087836 JAJ64972.1 JAN06901.1 KZ288347 PBC27523.1 GDIQ01116990 JAL34736.1 GDIP01016224 JAM87491.1 GDIP01221426 GDIQ01086149 JAJ01976.1 JAN08588.1 JR047615 AEY60459.1 GDIP01221425 JAJ01977.1 GDIQ01164620 JAK87105.1 GDIP01132934 JAL70780.1 GDIQ01100494 JAL51232.1 GDIP01186518 GDIQ01221345 LRGB01000626 JAJ36884.1 JAK30380.1 KZS17634.1 GDIP01239585 JAI83816.1 GDIQ01227590 GDIP01048247 JAK24135.1 JAM55468.1 PBC27522.1 GDIQ01245786 JAK05939.1 GDIP01088140 JAM15575.1 GDIQ01227589 GDIP01048246 JAK24136.1 JAM55469.1 GDIP01132933 JAL70781.1 GECZ01018907 JAS50862.1 GECU01026715 JAS80991.1 GDIQ01122776 JAL28950.1 KQ434786 KZC05224.1 KQ981281 KYN43279.1 GL888048 EGI68856.1 ABLF02014985 ADTU01027277 GFTR01007669 JAW08757.1 KQ978072 KYM97308.1 KQ982515 KYQ55578.1 GEDC01029473 JAS07825.1 GDKW01000536 JAI56059.1 JH431613 SAUD01029383 RXG54927.1 UFQS01001052 UFQT01001052 SSX08728.1 SSX28640.1 CM000070 KRT00518.1 EDY68141.1 QCYY01001784 ROT75338.1 OUUW01000007 SPP83114.1 GANO01002493 JAB57378.1 EF206694 ABQ00072.1 GAMC01008820 GAMC01008818 GAMC01008817 GAMC01008815 JAB97735.1 CH479179 EDW25095.1 RQTK01000245 RUS83385.1
RVE50559.1 KQ459602 KPI93385.1 KQ460615 KPJ13640.1 BABH01030221 JTDY01001556 KOB73544.1 LJIG01016384 KRT80762.1 ODYU01006090 SOQ47630.1 GBYB01003873 GBYB01013404 JAG73640.1 JAG83171.1 NEVH01000249 PNF43980.1 KK853131 KDR10924.1 GL732622 EFX70372.1 KQ762360 OAD55924.1 GEBQ01008313 JAT31664.1 KQ414681 KOC63923.1 GEBQ01010055 JAT29922.1 GDIQ01252053 JAJ99671.1 GDIQ01249041 JAK02684.1 GDIQ01250641 JAK01084.1 GDIP01112230 JAL91484.1 GDIP01045207 JAM58508.1 GDIP01160732 JAJ62670.1 GDIQ01026257 JAN68480.1 GDIQ01200394 JAK51331.1 GDIQ01252052 JAJ99672.1 GDIP01052899 JAM50816.1 GDIP01158430 GDIQ01087836 JAJ64972.1 JAN06901.1 KZ288347 PBC27523.1 GDIQ01116990 JAL34736.1 GDIP01016224 JAM87491.1 GDIP01221426 GDIQ01086149 JAJ01976.1 JAN08588.1 JR047615 AEY60459.1 GDIP01221425 JAJ01977.1 GDIQ01164620 JAK87105.1 GDIP01132934 JAL70780.1 GDIQ01100494 JAL51232.1 GDIP01186518 GDIQ01221345 LRGB01000626 JAJ36884.1 JAK30380.1 KZS17634.1 GDIP01239585 JAI83816.1 GDIQ01227590 GDIP01048247 JAK24135.1 JAM55468.1 PBC27522.1 GDIQ01245786 JAK05939.1 GDIP01088140 JAM15575.1 GDIQ01227589 GDIP01048246 JAK24136.1 JAM55469.1 GDIP01132933 JAL70781.1 GECZ01018907 JAS50862.1 GECU01026715 JAS80991.1 GDIQ01122776 JAL28950.1 KQ434786 KZC05224.1 KQ981281 KYN43279.1 GL888048 EGI68856.1 ABLF02014985 ADTU01027277 GFTR01007669 JAW08757.1 KQ978072 KYM97308.1 KQ982515 KYQ55578.1 GEDC01029473 JAS07825.1 GDKW01000536 JAI56059.1 JH431613 SAUD01029383 RXG54927.1 UFQS01001052 UFQT01001052 SSX08728.1 SSX28640.1 CM000070 KRT00518.1 EDY68141.1 QCYY01001784 ROT75338.1 OUUW01000007 SPP83114.1 GANO01002493 JAB57378.1 EF206694 ABQ00072.1 GAMC01008820 GAMC01008818 GAMC01008817 GAMC01008815 JAB97735.1 CH479179 EDW25095.1 RQTK01000245 RUS83385.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000053240
UP000005204
+ More
UP000037510 UP000235965 UP000027135 UP000000305 UP000053825 UP000242457 UP000005203 UP000076858 UP000076502 UP000078541 UP000007755 UP000007819 UP000005205 UP000078542 UP000075809 UP000002358 UP000288706 UP000091820 UP000001819 UP000283509 UP000268350 UP000078200 UP000008744 UP000271974
UP000037510 UP000235965 UP000027135 UP000000305 UP000053825 UP000242457 UP000005203 UP000076858 UP000076502 UP000078541 UP000007755 UP000007819 UP000005205 UP000078542 UP000075809 UP000002358 UP000288706 UP000091820 UP000001819 UP000283509 UP000268350 UP000078200 UP000008744 UP000271974
PRIDE
Interpro
IPR011545
DEAD/DEAH_box_helicase_dom
+ More
IPR014014 RNA_helicase_DEAD_Q_motif
IPR000629 RNA-helicase_DEAD-box_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR027417 P-loop_NTPase
IPR014722 Rib_L2_dom2
IPR023672 Ribosomal_L2_arc
IPR012340 NA-bd_OB-fold
IPR022669 Ribosomal_L2_C
IPR014726 Ribosomal_L2_dom3
IPR022666 Rbsml_prot_L2_RNA-bd_dom
IPR008991 Translation_prot_SH3-like_sf
IPR022671 Ribosomal_L2_CS
IPR002171 Ribosomal_L2
IPR014014 RNA_helicase_DEAD_Q_motif
IPR000629 RNA-helicase_DEAD-box_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR027417 P-loop_NTPase
IPR014722 Rib_L2_dom2
IPR023672 Ribosomal_L2_arc
IPR012340 NA-bd_OB-fold
IPR022669 Ribosomal_L2_C
IPR014726 Ribosomal_L2_dom3
IPR022666 Rbsml_prot_L2_RNA-bd_dom
IPR008991 Translation_prot_SH3-like_sf
IPR022671 Ribosomal_L2_CS
IPR002171 Ribosomal_L2
Gene 3D
CDD
ProteinModelPortal
A0A2A4K7F1
A0A2A4K6T1
A0A212EUG3
A0A437BJM2
A0A194PQH6
A0A194R8R4
+ More
H9JDJ7 A0A0L7LDT1 A0A0T6B0A8 A0A2H1W3F8 A0A0C9RAD5 A0A2J7RT20 A0A067R0G3 E9HD16 A0A310SAB9 A0A1B6M6Z0 A0A0L7QZA0 A0A1B6M1Y3 A0A0P5G8S5 A0A0P5G0W9 A0A0P5FWM3 A0A0P5UIY8 A0A0N8DD80 A0A0P5DLD9 A0A0P6I4V5 A0A0P5JG94 A0A0P5FSJ2 A0A0P5YZE3 A0A0N8DVH9 A0A2A3E6Y0 A0A0P5RNR6 A0A0P6BLR4 A0A0P6CZT9 V9IJF1 A0A0B4J2N5 A0A0N7ZQW4 A0A0P5MFB6 A0A0P5TAG6 A0A0P5S0Z4 A0A0P5IHA1 A0A0N7ZJE4 A0A0N8DC50 A0A2A3E8C1 A0A0P5G8K0 A0A0P5WC41 A0A0P5HM30 A0A0P5T0E3 A0A1B6FKZ8 A0A1B6I237 A0A0P5Q1M2 A0A154P1K4 A0A195FTR0 F4WAH3 J9K0U9 A0A158NVN2 A0A224X8R8 A0A195CB22 A0A151X5N7 A0A1B6C3L3 A0A0B4J2R6 A0A0P4W3R8 T1IWF1 A0A444SNF3 A0A1A9W9C9 A0A336MFD9 A0A0R3NP78 B5DWF0 A0A3R7PL42 A0A3B0JLC3 U5EU41 B0L0X1 A0A1A9VVY0 W8B905 B4G457 A0A433TP95
H9JDJ7 A0A0L7LDT1 A0A0T6B0A8 A0A2H1W3F8 A0A0C9RAD5 A0A2J7RT20 A0A067R0G3 E9HD16 A0A310SAB9 A0A1B6M6Z0 A0A0L7QZA0 A0A1B6M1Y3 A0A0P5G8S5 A0A0P5G0W9 A0A0P5FWM3 A0A0P5UIY8 A0A0N8DD80 A0A0P5DLD9 A0A0P6I4V5 A0A0P5JG94 A0A0P5FSJ2 A0A0P5YZE3 A0A0N8DVH9 A0A2A3E6Y0 A0A0P5RNR6 A0A0P6BLR4 A0A0P6CZT9 V9IJF1 A0A0B4J2N5 A0A0N7ZQW4 A0A0P5MFB6 A0A0P5TAG6 A0A0P5S0Z4 A0A0P5IHA1 A0A0N7ZJE4 A0A0N8DC50 A0A2A3E8C1 A0A0P5G8K0 A0A0P5WC41 A0A0P5HM30 A0A0P5T0E3 A0A1B6FKZ8 A0A1B6I237 A0A0P5Q1M2 A0A154P1K4 A0A195FTR0 F4WAH3 J9K0U9 A0A158NVN2 A0A224X8R8 A0A195CB22 A0A151X5N7 A0A1B6C3L3 A0A0B4J2R6 A0A0P4W3R8 T1IWF1 A0A444SNF3 A0A1A9W9C9 A0A336MFD9 A0A0R3NP78 B5DWF0 A0A3R7PL42 A0A3B0JLC3 U5EU41 B0L0X1 A0A1A9VVY0 W8B905 B4G457 A0A433TP95
PDB
6CZ5
E-value=2.25949e-179,
Score=1617
Ontologies
GO
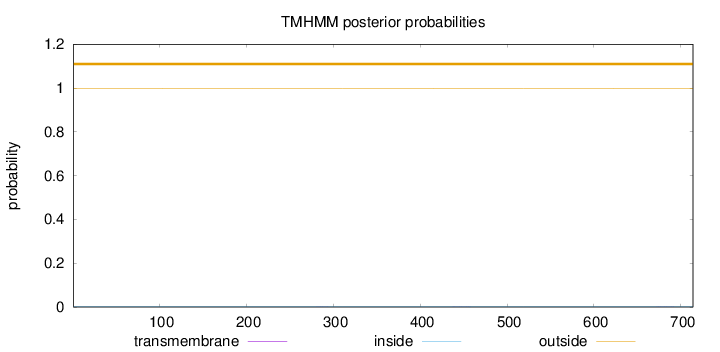
Topology
Length:
714
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0164
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00079
outside
1 - 714
Population Genetic Test Statistics
Pi
189.780415
Theta
178.75084
Tajima's D
-0.706363
CLR
288.188575
CSRT
0.195640217989101
Interpretation
Uncertain
