Pre Gene Modal
BGIBMGA007768
Annotation
PREDICTED:_UDP-glucuronic_acid_decarboxylase_1_isoform_X2_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.796 Mitochondrial Reliability : 1.331
Sequence
CDS
ATGAATTTGCAATCATCAAATGAAGTAACAAGTGATGATACAAAACATGAGCTTGCTGAAGCAAAAAATCGAATACAAGAATTAGAAAAAAAGATTGCTATTTTAGAAGGAAGAATGCCTCAAAAGTACCCTGAAGTAAAGTTTTTAGGATATAAAGAAAGGAGAAGAATCTTAGTGACAGGAGGAGCTGGTTTTGTTGGCTCACATTTAGTTGATATTCTCATGATGAAAGGTCATGAAGTCATTGTAGTCGACAATTTCTTTACTGGCCGGAAGAGAAACGTTGAGCATTGGTTCGGACATCGTAATTTTGAAATGATTCATCATGATATCGTCAATCCGTTGTATGTTGAAGCAGACGAAATATATCATTTAGCTAGTCCTGCCAGCCCACCGCATTATATGCAAAATCCTGTTAAAACAATCAAGACTAATACTCTAGGAACTATAAATATGTTAGGTCTCGCCAGACGCGTCGGCGCTAGAATACTGATCGCAAGCACTTCAGAAGTATATGGAGATCCTTTGGTGCATCCACAACCTGAATCTTATTGGGGCCATGTTAATCCAATTGGCCCTCGAGCGTGTTACGACGAAGGCAAGAGAGTTGCAGAGACACTGGCGTATTCGTACGCAAAACAAGAGAAAGTCGACGTGAGAGTCGCTAGGATATTCAACACGTACGGACCTAGAATGCATGTTTCAGACGGTCGTGTAGTTTCTAATTTCATAATGCAAGCTTTACAGAATTTAACTATTACAGTTTATGGTAACGGAAAACAGACCCGATCGTTCTGCTACGTGTCTGACCTTGTCGATGGACTGCTAGGCTTAATGGCTTCTAGTTATACATTACCTGTTAACATTGGGAACCCGGTAGAACATACTATAGAAGAATTTGCAATGATCATAAAGAATCTAGTCCCCGGGTGCCGCAGTACTGTCGCCACGGGAGCCGCGGTCGAGGACGACCCCCAGCGGCGCCGACCGGACATTGCAGTGGCGGAGGCCCATTTACACTGGAAACCTAAAGTAAGCCTACAGGAAGGCTTGCAAAGAACCATCGACTATTTCAAAGAAGAATTATCAAGGACTACGCTTTACAATAATCAAACTTATGTAGATATGAAAGTCAAACATCATTAG
Protein
MNLQSSNEVTSDDTKHELAEAKNRIQELEKKIAILEGRMPQKYPEVKFLGYKERRRILVTGGAGFVGSHLVDILMMKGHEVIVVDNFFTGRKRNVEHWFGHRNFEMIHHDIVNPLYVEADEIYHLASPASPPHYMQNPVKTIKTNTLGTINMLGLARRVGARILIASTSEVYGDPLVHPQPESYWGHVNPIGPRACYDEGKRVAETLAYSYAKQEKVDVRVARIFNTYGPRMHVSDGRVVSNFIMQALQNLTITVYGNGKQTRSFCYVSDLVDGLLGLMASSYTLPVNIGNPVEHTIEEFAMIIKNLVPGCRSTVATGAAVEDDPQRRRPDIAVAEAHLHWKPKVSLQEGLQRTIDYFKEELSRTTLYNNQTYVDMKVKHH
Summary
Uniprot
H9JE21
A0A3S2LNM3
A0A2H1VJ71
A0A2A4K736
A0A212EUI2
A0A194PK32
+ More
A0A182FPF6 A0A182L704 A0A182RX71 W5JFY7 A0A182PS67 A0A2M4CHL1 A0A182MG10 A0A2M3Z770 A0A182ID60 A0A2M4AIC8 A0A2M4AI85 Q7QAZ6 A0A182XG14 A0A182XX25 A0A2M4AID7 A0A2M4AIS5 A0A2M4BLF8 A0A2M4BLI6 A0A2M4BM40 A0A182TUH9 A0A1Q3G2J8 A0A182W174 A0A182VIX4 A0A2P8YU34 A0A1L8E2D9 A0A182NCX1 A0A1L8E1X4 A0A182Q1I4 A0A2J7RJ47 A0A1B0C9X2 A0A182JUK5 A0A182JET1 A0A023ETQ3 Q176W8 A0A084WP06 A0A067QXX8 Q176W9 D7EL37 A0A0K8WGL0 A0A1W4WU98 A0A034VY56 A0A232F1Y6 W8BUR8 E9GL34 A0A0L0BS88 K7IQZ2 B3M9M4 U5EXQ8 B4J221 A0A0P5NAB9 E2AEW9 A0A0P5SNP8 B4PCU0 B4HJD6 A0A0J7L078 Q9VSE8 B4QLJ2 E2BR94 T1PHL1 A0A1I8NMN5 A0A1I8NMN0 B4LCC0 A0A026WW16 B4L927 B4N4C6 B3NF42 A0A1B0FKT8 A0A1A9UHB3 A0A1B0BMW8 A0A1A9ZSB8 A0A3B0KTN6 B4H177 A0A2A3EIT7 A0A088AAG1 A0A1A9XD44 Q29FJ1 A0A1W4UFC2 A0A0M4EKY4 A0A1A9WGG9 T1I6Q9 A0A0L7R4K8 F4W8W3 A0A3L8D9E9 A0A1Y1LYX0 A0A151WZN4 A0A1B6DQ87 A0A195CVA6 A0A1D2NIN6 A0A158NHH4 A0A195BVY3 J3JUJ3 A0A2J7RJ48 A0A1J1I9I5 A0A2S2Q916
A0A182FPF6 A0A182L704 A0A182RX71 W5JFY7 A0A182PS67 A0A2M4CHL1 A0A182MG10 A0A2M3Z770 A0A182ID60 A0A2M4AIC8 A0A2M4AI85 Q7QAZ6 A0A182XG14 A0A182XX25 A0A2M4AID7 A0A2M4AIS5 A0A2M4BLF8 A0A2M4BLI6 A0A2M4BM40 A0A182TUH9 A0A1Q3G2J8 A0A182W174 A0A182VIX4 A0A2P8YU34 A0A1L8E2D9 A0A182NCX1 A0A1L8E1X4 A0A182Q1I4 A0A2J7RJ47 A0A1B0C9X2 A0A182JUK5 A0A182JET1 A0A023ETQ3 Q176W8 A0A084WP06 A0A067QXX8 Q176W9 D7EL37 A0A0K8WGL0 A0A1W4WU98 A0A034VY56 A0A232F1Y6 W8BUR8 E9GL34 A0A0L0BS88 K7IQZ2 B3M9M4 U5EXQ8 B4J221 A0A0P5NAB9 E2AEW9 A0A0P5SNP8 B4PCU0 B4HJD6 A0A0J7L078 Q9VSE8 B4QLJ2 E2BR94 T1PHL1 A0A1I8NMN5 A0A1I8NMN0 B4LCC0 A0A026WW16 B4L927 B4N4C6 B3NF42 A0A1B0FKT8 A0A1A9UHB3 A0A1B0BMW8 A0A1A9ZSB8 A0A3B0KTN6 B4H177 A0A2A3EIT7 A0A088AAG1 A0A1A9XD44 Q29FJ1 A0A1W4UFC2 A0A0M4EKY4 A0A1A9WGG9 T1I6Q9 A0A0L7R4K8 F4W8W3 A0A3L8D9E9 A0A1Y1LYX0 A0A151WZN4 A0A1B6DQ87 A0A195CVA6 A0A1D2NIN6 A0A158NHH4 A0A195BVY3 J3JUJ3 A0A2J7RJ48 A0A1J1I9I5 A0A2S2Q916
Pubmed
19121390
22118469
26354079
20966253
20920257
23761445
+ More
12364791 14747013 17210077 25244985 29403074 24945155 17510324 24438588 24845553 18362917 19820115 25348373 28648823 24495485 21292972 26108605 20075255 17994087 20798317 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25315136 24508170 30249741 15632085 21719571 28004739 27289101 21347285 22516182 23537049
12364791 14747013 17210077 25244985 29403074 24945155 17510324 24438588 24845553 18362917 19820115 25348373 28648823 24495485 21292972 26108605 20075255 17994087 20798317 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25315136 24508170 30249741 15632085 21719571 28004739 27289101 21347285 22516182 23537049
EMBL
BABH01002483
BABH01002484
RSAL01000046
RVE50564.1
ODYU01002850
SOQ40846.1
+ More
NWSH01000076 PCG79889.1 AGBW02012389 OWR45143.1 KQ459602 KPI93378.1 ADMH02001267 ETN63287.1 GGFL01000615 MBW64793.1 AXCM01006078 GGFM01003603 MBW24354.1 APCN01005034 GGFK01007176 MBW40497.1 GGFK01007175 MBW40496.1 AAAB01008880 EAA08612.5 GGFK01007186 MBW40507.1 GGFK01007187 MBW40508.1 GGFJ01004769 MBW53910.1 GGFJ01004786 MBW53927.1 GGFJ01004872 MBW54013.1 GFDL01001022 JAV34023.1 PYGN01000359 PSN47752.1 GFDF01001409 JAV12675.1 GFDF01001410 JAV12674.1 AXCN02001559 NEVH01003007 PNF40854.1 AJWK01002981 GAPW01001804 JAC11794.1 CH477381 EAT42204.1 ATLV01024805 KE525361 KFB51950.1 KK853225 KDR09682.1 EAT42203.1 DS497802 EFA11835.1 GDHF01003204 GDHF01002105 JAI49110.1 JAI50209.1 GAKP01012159 GAKP01012157 GAKP01012154 GAKP01012152 JAC46795.1 NNAY01001282 OXU24499.1 GAMC01001455 JAC05101.1 GL732550 EFX79862.1 JRES01001437 KNC22930.1 CH902618 EDV39030.1 GANO01002424 JAB57447.1 CH916366 EDV95946.1 GDIQ01151929 JAK99796.1 GL438952 EFN68014.1 GDIQ01206666 GDIP01140896 JAK45059.1 JAL62818.1 CM000159 EDW93844.1 CH480815 EDW40660.1 LBMM01001601 KMQ96036.1 AE014296 AY051913 AAF50474.1 AAK93337.1 CM000363 CM002912 EDX09652.1 KMY98299.1 GL449936 EFN81783.1 KA648256 AFP62885.1 CH940647 EDW68765.1 KK107078 QOIP01000011 EZA60192.1 RLU16742.1 CH933816 EDW17202.1 CH964095 EDW79000.2 CH954178 EDV50384.1 CCAG010001046 JXJN01017069 OUUW01000012 SPP87258.1 CH479202 EDW30054.1 KZ288236 PBC31384.1 CH379067 EAL31263.2 CP012525 ALC44926.1 ACPB03010138 ACPB03010139 KQ414657 KOC65696.1 GL887973 EGI69355.1 RLU16772.1 GEZM01046440 JAV77235.1 KQ982648 KYQ53227.1 GEDC01009467 JAS27831.1 KQ977259 KYN04565.1 LJIJ01000032 ODN04975.1 ADTU01001682 KQ976398 KYM92742.1 APGK01037400 BT126907 KB740948 AEE61869.1 ENN77320.1 PNF40856.1 CVRI01000045 CRK96943.1 GGMS01004857 MBY74060.1
NWSH01000076 PCG79889.1 AGBW02012389 OWR45143.1 KQ459602 KPI93378.1 ADMH02001267 ETN63287.1 GGFL01000615 MBW64793.1 AXCM01006078 GGFM01003603 MBW24354.1 APCN01005034 GGFK01007176 MBW40497.1 GGFK01007175 MBW40496.1 AAAB01008880 EAA08612.5 GGFK01007186 MBW40507.1 GGFK01007187 MBW40508.1 GGFJ01004769 MBW53910.1 GGFJ01004786 MBW53927.1 GGFJ01004872 MBW54013.1 GFDL01001022 JAV34023.1 PYGN01000359 PSN47752.1 GFDF01001409 JAV12675.1 GFDF01001410 JAV12674.1 AXCN02001559 NEVH01003007 PNF40854.1 AJWK01002981 GAPW01001804 JAC11794.1 CH477381 EAT42204.1 ATLV01024805 KE525361 KFB51950.1 KK853225 KDR09682.1 EAT42203.1 DS497802 EFA11835.1 GDHF01003204 GDHF01002105 JAI49110.1 JAI50209.1 GAKP01012159 GAKP01012157 GAKP01012154 GAKP01012152 JAC46795.1 NNAY01001282 OXU24499.1 GAMC01001455 JAC05101.1 GL732550 EFX79862.1 JRES01001437 KNC22930.1 CH902618 EDV39030.1 GANO01002424 JAB57447.1 CH916366 EDV95946.1 GDIQ01151929 JAK99796.1 GL438952 EFN68014.1 GDIQ01206666 GDIP01140896 JAK45059.1 JAL62818.1 CM000159 EDW93844.1 CH480815 EDW40660.1 LBMM01001601 KMQ96036.1 AE014296 AY051913 AAF50474.1 AAK93337.1 CM000363 CM002912 EDX09652.1 KMY98299.1 GL449936 EFN81783.1 KA648256 AFP62885.1 CH940647 EDW68765.1 KK107078 QOIP01000011 EZA60192.1 RLU16742.1 CH933816 EDW17202.1 CH964095 EDW79000.2 CH954178 EDV50384.1 CCAG010001046 JXJN01017069 OUUW01000012 SPP87258.1 CH479202 EDW30054.1 KZ288236 PBC31384.1 CH379067 EAL31263.2 CP012525 ALC44926.1 ACPB03010138 ACPB03010139 KQ414657 KOC65696.1 GL887973 EGI69355.1 RLU16772.1 GEZM01046440 JAV77235.1 KQ982648 KYQ53227.1 GEDC01009467 JAS27831.1 KQ977259 KYN04565.1 LJIJ01000032 ODN04975.1 ADTU01001682 KQ976398 KYM92742.1 APGK01037400 BT126907 KB740948 AEE61869.1 ENN77320.1 PNF40856.1 CVRI01000045 CRK96943.1 GGMS01004857 MBY74060.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000069272
+ More
UP000075882 UP000075900 UP000000673 UP000075885 UP000075883 UP000075840 UP000007062 UP000076407 UP000076408 UP000075902 UP000075920 UP000075903 UP000245037 UP000075884 UP000075886 UP000235965 UP000092461 UP000075881 UP000075880 UP000008820 UP000030765 UP000027135 UP000007266 UP000192223 UP000215335 UP000000305 UP000037069 UP000002358 UP000007801 UP000001070 UP000000311 UP000002282 UP000001292 UP000036403 UP000000803 UP000000304 UP000008237 UP000095301 UP000095300 UP000008792 UP000053097 UP000279307 UP000009192 UP000007798 UP000008711 UP000092444 UP000078200 UP000092460 UP000092445 UP000268350 UP000008744 UP000242457 UP000005203 UP000092443 UP000001819 UP000192221 UP000092553 UP000091820 UP000015103 UP000053825 UP000007755 UP000075809 UP000078542 UP000094527 UP000005205 UP000078540 UP000019118 UP000183832
UP000075882 UP000075900 UP000000673 UP000075885 UP000075883 UP000075840 UP000007062 UP000076407 UP000076408 UP000075902 UP000075920 UP000075903 UP000245037 UP000075884 UP000075886 UP000235965 UP000092461 UP000075881 UP000075880 UP000008820 UP000030765 UP000027135 UP000007266 UP000192223 UP000215335 UP000000305 UP000037069 UP000002358 UP000007801 UP000001070 UP000000311 UP000002282 UP000001292 UP000036403 UP000000803 UP000000304 UP000008237 UP000095301 UP000095300 UP000008792 UP000053097 UP000279307 UP000009192 UP000007798 UP000008711 UP000092444 UP000078200 UP000092460 UP000092445 UP000268350 UP000008744 UP000242457 UP000005203 UP000092443 UP000001819 UP000192221 UP000092553 UP000091820 UP000015103 UP000053825 UP000007755 UP000075809 UP000078542 UP000094527 UP000005205 UP000078540 UP000019118 UP000183832
Pfam
Interpro
IPR036291
NAD(P)-bd_dom_sf
+ More
IPR016040 NAD(P)-bd_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR036640 ABC1_TM_sf
IPR027417 P-loop_NTPase
IPR011527 ABC1_TM_dom
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR003593 AAA+_ATPase
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR021977 Beta-TrCP_D
IPR001377 Ribosomal_S6e
IPR001810 F-box_dom
IPR017986 WD40_repeat_dom
IPR018282 Ribosomal_S6e_CS
IPR001680 WD40_repeat
IPR036047 F-box-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR016040 NAD(P)-bd_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR036640 ABC1_TM_sf
IPR027417 P-loop_NTPase
IPR011527 ABC1_TM_dom
IPR017871 ABC_transporter_CS
IPR003439 ABC_transporter-like
IPR003593 AAA+_ATPase
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR012934 Znf_AD
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR021977 Beta-TrCP_D
IPR001377 Ribosomal_S6e
IPR001810 F-box_dom
IPR017986 WD40_repeat_dom
IPR018282 Ribosomal_S6e_CS
IPR001680 WD40_repeat
IPR036047 F-box-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JE21
A0A3S2LNM3
A0A2H1VJ71
A0A2A4K736
A0A212EUI2
A0A194PK32
+ More
A0A182FPF6 A0A182L704 A0A182RX71 W5JFY7 A0A182PS67 A0A2M4CHL1 A0A182MG10 A0A2M3Z770 A0A182ID60 A0A2M4AIC8 A0A2M4AI85 Q7QAZ6 A0A182XG14 A0A182XX25 A0A2M4AID7 A0A2M4AIS5 A0A2M4BLF8 A0A2M4BLI6 A0A2M4BM40 A0A182TUH9 A0A1Q3G2J8 A0A182W174 A0A182VIX4 A0A2P8YU34 A0A1L8E2D9 A0A182NCX1 A0A1L8E1X4 A0A182Q1I4 A0A2J7RJ47 A0A1B0C9X2 A0A182JUK5 A0A182JET1 A0A023ETQ3 Q176W8 A0A084WP06 A0A067QXX8 Q176W9 D7EL37 A0A0K8WGL0 A0A1W4WU98 A0A034VY56 A0A232F1Y6 W8BUR8 E9GL34 A0A0L0BS88 K7IQZ2 B3M9M4 U5EXQ8 B4J221 A0A0P5NAB9 E2AEW9 A0A0P5SNP8 B4PCU0 B4HJD6 A0A0J7L078 Q9VSE8 B4QLJ2 E2BR94 T1PHL1 A0A1I8NMN5 A0A1I8NMN0 B4LCC0 A0A026WW16 B4L927 B4N4C6 B3NF42 A0A1B0FKT8 A0A1A9UHB3 A0A1B0BMW8 A0A1A9ZSB8 A0A3B0KTN6 B4H177 A0A2A3EIT7 A0A088AAG1 A0A1A9XD44 Q29FJ1 A0A1W4UFC2 A0A0M4EKY4 A0A1A9WGG9 T1I6Q9 A0A0L7R4K8 F4W8W3 A0A3L8D9E9 A0A1Y1LYX0 A0A151WZN4 A0A1B6DQ87 A0A195CVA6 A0A1D2NIN6 A0A158NHH4 A0A195BVY3 J3JUJ3 A0A2J7RJ48 A0A1J1I9I5 A0A2S2Q916
A0A182FPF6 A0A182L704 A0A182RX71 W5JFY7 A0A182PS67 A0A2M4CHL1 A0A182MG10 A0A2M3Z770 A0A182ID60 A0A2M4AIC8 A0A2M4AI85 Q7QAZ6 A0A182XG14 A0A182XX25 A0A2M4AID7 A0A2M4AIS5 A0A2M4BLF8 A0A2M4BLI6 A0A2M4BM40 A0A182TUH9 A0A1Q3G2J8 A0A182W174 A0A182VIX4 A0A2P8YU34 A0A1L8E2D9 A0A182NCX1 A0A1L8E1X4 A0A182Q1I4 A0A2J7RJ47 A0A1B0C9X2 A0A182JUK5 A0A182JET1 A0A023ETQ3 Q176W8 A0A084WP06 A0A067QXX8 Q176W9 D7EL37 A0A0K8WGL0 A0A1W4WU98 A0A034VY56 A0A232F1Y6 W8BUR8 E9GL34 A0A0L0BS88 K7IQZ2 B3M9M4 U5EXQ8 B4J221 A0A0P5NAB9 E2AEW9 A0A0P5SNP8 B4PCU0 B4HJD6 A0A0J7L078 Q9VSE8 B4QLJ2 E2BR94 T1PHL1 A0A1I8NMN5 A0A1I8NMN0 B4LCC0 A0A026WW16 B4L927 B4N4C6 B3NF42 A0A1B0FKT8 A0A1A9UHB3 A0A1B0BMW8 A0A1A9ZSB8 A0A3B0KTN6 B4H177 A0A2A3EIT7 A0A088AAG1 A0A1A9XD44 Q29FJ1 A0A1W4UFC2 A0A0M4EKY4 A0A1A9WGG9 T1I6Q9 A0A0L7R4K8 F4W8W3 A0A3L8D9E9 A0A1Y1LYX0 A0A151WZN4 A0A1B6DQ87 A0A195CVA6 A0A1D2NIN6 A0A158NHH4 A0A195BVY3 J3JUJ3 A0A2J7RJ48 A0A1J1I9I5 A0A2S2Q916
PDB
2B69
E-value=1.07368e-134,
Score=1229
Ontologies
PATHWAY
GO
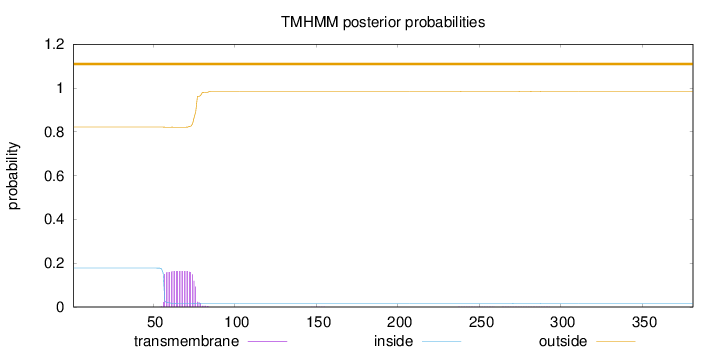
Topology
Length:
381
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.25234000000001
Exp number, first 60 AAs:
0.65298
Total prob of N-in:
0.17787
outside
1 - 381
Population Genetic Test Statistics
Pi
205.909226
Theta
163.455633
Tajima's D
0.895529
CLR
0.00465
CSRT
0.630818459077046
Interpretation
Uncertain
