Gene
KWMTBOMO09028
Pre Gene Modal
BGIBMGA007765
Annotation
PREDICTED:_beta-1?3-galactosyltransferase_6_[Papilio_polytes]
Full name
Hexosyltransferase
Location in the cell
Cytoplasmic Reliability : 1.633 Extracellular Reliability : 1.005 Nuclear Reliability : 1.417
Sequence
CDS
ATGTGCAGAAGTAATGATATGCTTTTAATTGATGATCTCCAAGATTCCTATGATAATTTAACATCCAAAGTATTAAAAGCATTGAAGTGGTTTAGTGAAAATTTAAAAAATCTTCGATACTTGATAAAATGTGATGATGATTCATTTGTTAGAATAGATTTGATAGTGAGAGATCTGGAAGCATTTGCTCCGAAAATGGATGGAGAAGAAATAAAATCATTTGTTTCGTATAAGGTTGGTTGGTTGAAATCTCATTCTGCATATAATGGATTATATTGGGGATATTTCAGTGGAAAAGCCAAAGTGTTTATGTCTGGCAAGTGGAAAGAGACAGATTGGTTCCTTTGTGATAACTACTTGCCTTATGCTCGTGGTGGAGGATATGTTATATCTCGTAGCATAGTCGATTTCGTTGGTAAAAATGCTGAATTATTGAGTCATTACAATTCTGAAGATGTGTCAATGGGAGTGTGGACAGCACCATTAGACAGAATCAATAGGGTTCATGATGTCAGATTTGACACTGAATGGAAGACTAGAGGTTGCTTGACAAATATGTTAGTTCGACATAAACAAACCCCAAGTGATATGTTTCAGTTATTTAAAAATTTGATTAATTCACATGGCAATAAACTTTGCAAACATGAAAGTACGGAAAGGAAATCATATCATTACAACTGGAATGTTCTTCCTAGTGAATGCTGTCACTAA
Protein
MCRSNDMLLIDDLQDSYDNLTSKVLKALKWFSENLKNLRYLIKCDDDSFVRIDLIVRDLEAFAPKMDGEEIKSFVSYKVGWLKSHSAYNGLYWGYFSGKAKVFMSGKWKETDWFLCDNYLPYARGGGYVISRSIVDFVGKNAELLSHYNSEDVSMGVWTAPLDRINRVHDVRFDTEWKTRGCLTNMLVRHKQTPSDMFQLFKNLINSHGNKLCKHESTERKSYHYNWNVLPSECCH
Summary
Similarity
Belongs to the glycosyltransferase 31 family.
Feature
chain Hexosyltransferase
Uniprot
H9JE18
A0A2H1VJ31
A0A2A4K6M7
A0A212EUF8
S4NXU0
A0A437AT78
+ More
A0A437BJX4 A0A194PKX0 A0A0L7LEZ7 A0A194R736 A0A1B6LE51 A0A443QFV8 A0A0P5AJC0 A0A3S4Q954 A0A0P4YW08 A0A0N7ZQZ9 A0A0P5XMH1 A0A0P5J703 A0A0N8D9I0 A0A0P5LSB6 A0A0P5Q469 A0A0P5GUR7 A0A0P5ZZZ2 A0A0P5HLV2 A0A1B6I698 A0A0P5R7P5 A0A1Y3B2A5 E9HTR4 A0A1S3DAY2 A0A0P5RGA7 A0A0P6HVR1 A0A099ZBT2 A0A0P5V9Y5 A0A0P5WXH4 A0A2B4SVP5 A0A067RFU4 A0A1B6EPG0 A0A433TIW4 A0A2I0M2T7 A0A158P1A5 A0A2T7PQF8 A0A195DL48 F4WA33 A0A151I5W5 A0A151IG30 A0A2J7QK23 A0A091HXN7 A0A093GPU1 A0A091NFV3 A0A151XH51 E9IKJ2 A0A1V4KNU9 A0A151K018 U3KJK4 A0A091IM70 G3UU25 A0A091LG01 A0A091F8A8 A0A091RQ55 A0A091KDC8 A0A091JNK1 A0A0T6AWB4 A0A093C9C9 A0A3M0ISV9 A0A091RBA2 H0W6A7 H0Z0U9 A0A091FV83 A0A2P4TG69 A0A250XYI0 A0A091NSA0 A0A226N0L9 A0A091UZY6 A0A0A0AV72 A0A2I0TS58 A0A087VRW8 A0A093IXD0 A0A093H085 A0A093NUM9 A0A1B6D0R3 A0A3L8Q8V4 A0A087QSA9 A0A091US57 A0A093ISC4 A0A226PDQ6 A0A094L8X7 A0A087ZNU1 A0A0J7KDG7 K7EYM3 U3I5J1 E2ATY8 A0A132A8J8 A7SZM8 A0A0L7RAS5 A0A154PA43 A0A094KRT1 A0A232F6A2 A0A091UIZ1 K7JRP2 A0A091WLH4
A0A437BJX4 A0A194PKX0 A0A0L7LEZ7 A0A194R736 A0A1B6LE51 A0A443QFV8 A0A0P5AJC0 A0A3S4Q954 A0A0P4YW08 A0A0N7ZQZ9 A0A0P5XMH1 A0A0P5J703 A0A0N8D9I0 A0A0P5LSB6 A0A0P5Q469 A0A0P5GUR7 A0A0P5ZZZ2 A0A0P5HLV2 A0A1B6I698 A0A0P5R7P5 A0A1Y3B2A5 E9HTR4 A0A1S3DAY2 A0A0P5RGA7 A0A0P6HVR1 A0A099ZBT2 A0A0P5V9Y5 A0A0P5WXH4 A0A2B4SVP5 A0A067RFU4 A0A1B6EPG0 A0A433TIW4 A0A2I0M2T7 A0A158P1A5 A0A2T7PQF8 A0A195DL48 F4WA33 A0A151I5W5 A0A151IG30 A0A2J7QK23 A0A091HXN7 A0A093GPU1 A0A091NFV3 A0A151XH51 E9IKJ2 A0A1V4KNU9 A0A151K018 U3KJK4 A0A091IM70 G3UU25 A0A091LG01 A0A091F8A8 A0A091RQ55 A0A091KDC8 A0A091JNK1 A0A0T6AWB4 A0A093C9C9 A0A3M0ISV9 A0A091RBA2 H0W6A7 H0Z0U9 A0A091FV83 A0A2P4TG69 A0A250XYI0 A0A091NSA0 A0A226N0L9 A0A091UZY6 A0A0A0AV72 A0A2I0TS58 A0A087VRW8 A0A093IXD0 A0A093H085 A0A093NUM9 A0A1B6D0R3 A0A3L8Q8V4 A0A087QSA9 A0A091US57 A0A093ISC4 A0A226PDQ6 A0A094L8X7 A0A087ZNU1 A0A0J7KDG7 K7EYM3 U3I5J1 E2ATY8 A0A132A8J8 A7SZM8 A0A0L7RAS5 A0A154PA43 A0A094KRT1 A0A232F6A2 A0A091UIZ1 K7JRP2 A0A091WLH4
EC Number
2.4.1.-
Pubmed
EMBL
BABH01002484
ODYU01002850
SOQ40845.1
NWSH01000076
PCG79891.1
AGBW02012389
+ More
OWR45142.1 GAIX01012022 JAA80538.1 RSAL01001151 RVE40918.1 RSAL01000046 RVE50565.1 KQ459602 KPI93379.1 JTDY01001406 KOB73954.1 KQ460615 KPJ13633.1 GEBQ01018028 JAT21949.1 NCKU01008426 RWS01903.1 GDIP01198499 JAJ24903.1 NCKU01011423 RWS00446.1 GDIP01222301 JAJ01101.1 GDIP01221145 JAJ02257.1 GDIP01072238 LRGB01002849 JAM31477.1 KZS05998.1 GDIQ01204311 JAK47414.1 GDIP01055607 JAM48108.1 GDIQ01165983 JAK85742.1 GDIQ01126377 JAL25349.1 GDIQ01242310 JAK09415.1 GDIP01049517 JAM54198.1 GDIQ01248492 GDIQ01030239 JAK03233.1 GECU01025255 JAS82451.1 GDIQ01109589 JAL42137.1 MUJZ01044537 OTF74941.1 GL732783 EFX64867.1 GDIQ01107442 JAL44284.1 GDIQ01038082 JAN56655.1 KL891033 KGL78305.1 GDIP01103394 JAM00321.1 GDIP01080150 JAM23565.1 LSMT01000022 PFX32537.1 KK852714 KDR17902.1 GECZ01029953 JAS39816.1 RQTK01000334 RUS81504.1 AKCR02000044 PKK23992.1 ADTU01006170 PZQS01000002 PVD35607.1 KQ980762 KYN13571.1 GL888037 EGI68963.1 KQ976402 KYM92287.1 KQ977743 KYN00172.1 NEVH01013361 PNF28926.1 KL528423 KFO91983.1 KL216962 KFV72323.1 KK846972 KFP88361.1 KQ982138 KYQ59687.1 GL763991 EFZ18894.1 LSYS01002427 OPJ86091.1 KQ981301 KYN43054.1 AGTO01012312 KL218792 KFP08420.1 KK751327 KFP41861.1 KK719670 KFO64809.1 KK928829 KFQ44209.1 KK553267 KFP34396.1 KK502305 KFP22544.1 LJIG01022650 KRT79443.1 KL456461 KFV09584.1 QRBI01000235 RMB91602.1 KK715653 KFQ36209.1 AAKN02055863 ABQF01059082 KL447398 KFO73109.1 PPHD01000532 POI35372.1 GFFV01003545 JAV36400.1 KL392719 KFP92201.1 MCFN01000301 OXB60998.1 KK407696 KFQ83113.1 KL872854 KGL96955.1 KZ507538 PKU36629.1 KL503292 KFO15360.1 KK564434 KFW03414.1 KK614945 KFV48143.1 KL225141 KFW67896.1 GEDC01018027 GEDC01017675 JAS19271.1 JAS19623.1 QUSF01002021 QUSF01002020 QUSF01002019 QUSF01002018 RLV63754.1 RLV63755.1 RLV63756.1 RLV63757.1 KL225853 KFM04113.1 KL410063 KFQ93784.1 KK594876 KFW02517.1 AWGT02000089 OXB78175.1 KL282093 KFZ67778.1 LBMM01009298 KMQ88236.1 AGCU01113407 ADON01000017 ADON01000018 ADON01000019 GL442747 EFN63119.1 JXLN01011455 KPM07284.1 DS469975 EDO30842.1 KQ414618 KOC67943.1 KQ434857 KZC08806.1 KL341666 KFZ53277.1 NNAY01000815 OXU26361.1 KK453906 KFQ75225.1 KK735901 KFR16409.1
OWR45142.1 GAIX01012022 JAA80538.1 RSAL01001151 RVE40918.1 RSAL01000046 RVE50565.1 KQ459602 KPI93379.1 JTDY01001406 KOB73954.1 KQ460615 KPJ13633.1 GEBQ01018028 JAT21949.1 NCKU01008426 RWS01903.1 GDIP01198499 JAJ24903.1 NCKU01011423 RWS00446.1 GDIP01222301 JAJ01101.1 GDIP01221145 JAJ02257.1 GDIP01072238 LRGB01002849 JAM31477.1 KZS05998.1 GDIQ01204311 JAK47414.1 GDIP01055607 JAM48108.1 GDIQ01165983 JAK85742.1 GDIQ01126377 JAL25349.1 GDIQ01242310 JAK09415.1 GDIP01049517 JAM54198.1 GDIQ01248492 GDIQ01030239 JAK03233.1 GECU01025255 JAS82451.1 GDIQ01109589 JAL42137.1 MUJZ01044537 OTF74941.1 GL732783 EFX64867.1 GDIQ01107442 JAL44284.1 GDIQ01038082 JAN56655.1 KL891033 KGL78305.1 GDIP01103394 JAM00321.1 GDIP01080150 JAM23565.1 LSMT01000022 PFX32537.1 KK852714 KDR17902.1 GECZ01029953 JAS39816.1 RQTK01000334 RUS81504.1 AKCR02000044 PKK23992.1 ADTU01006170 PZQS01000002 PVD35607.1 KQ980762 KYN13571.1 GL888037 EGI68963.1 KQ976402 KYM92287.1 KQ977743 KYN00172.1 NEVH01013361 PNF28926.1 KL528423 KFO91983.1 KL216962 KFV72323.1 KK846972 KFP88361.1 KQ982138 KYQ59687.1 GL763991 EFZ18894.1 LSYS01002427 OPJ86091.1 KQ981301 KYN43054.1 AGTO01012312 KL218792 KFP08420.1 KK751327 KFP41861.1 KK719670 KFO64809.1 KK928829 KFQ44209.1 KK553267 KFP34396.1 KK502305 KFP22544.1 LJIG01022650 KRT79443.1 KL456461 KFV09584.1 QRBI01000235 RMB91602.1 KK715653 KFQ36209.1 AAKN02055863 ABQF01059082 KL447398 KFO73109.1 PPHD01000532 POI35372.1 GFFV01003545 JAV36400.1 KL392719 KFP92201.1 MCFN01000301 OXB60998.1 KK407696 KFQ83113.1 KL872854 KGL96955.1 KZ507538 PKU36629.1 KL503292 KFO15360.1 KK564434 KFW03414.1 KK614945 KFV48143.1 KL225141 KFW67896.1 GEDC01018027 GEDC01017675 JAS19271.1 JAS19623.1 QUSF01002021 QUSF01002020 QUSF01002019 QUSF01002018 RLV63754.1 RLV63755.1 RLV63756.1 RLV63757.1 KL225853 KFM04113.1 KL410063 KFQ93784.1 KK594876 KFW02517.1 AWGT02000089 OXB78175.1 KL282093 KFZ67778.1 LBMM01009298 KMQ88236.1 AGCU01113407 ADON01000017 ADON01000018 ADON01000019 GL442747 EFN63119.1 JXLN01011455 KPM07284.1 DS469975 EDO30842.1 KQ414618 KOC67943.1 KQ434857 KZC08806.1 KL341666 KFZ53277.1 NNAY01000815 OXU26361.1 KK453906 KFQ75225.1 KK735901 KFR16409.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000037510
+ More
UP000053240 UP000285301 UP000076858 UP000000305 UP000079169 UP000053641 UP000225706 UP000027135 UP000271974 UP000053872 UP000005205 UP000245119 UP000078492 UP000007755 UP000078540 UP000078542 UP000235965 UP000053875 UP000075809 UP000190648 UP000078541 UP000016665 UP000054308 UP000001645 UP000052976 UP000053119 UP000269221 UP000005447 UP000007754 UP000053760 UP000198323 UP000053858 UP000054081 UP000276834 UP000053286 UP000053283 UP000198419 UP000005203 UP000036403 UP000007267 UP000016666 UP000000311 UP000001593 UP000053825 UP000076502 UP000215335 UP000002358 UP000053605
UP000053240 UP000285301 UP000076858 UP000000305 UP000079169 UP000053641 UP000225706 UP000027135 UP000271974 UP000053872 UP000005205 UP000245119 UP000078492 UP000007755 UP000078540 UP000078542 UP000235965 UP000053875 UP000075809 UP000190648 UP000078541 UP000016665 UP000054308 UP000001645 UP000052976 UP000053119 UP000269221 UP000005447 UP000007754 UP000053760 UP000198323 UP000053858 UP000054081 UP000276834 UP000053286 UP000053283 UP000198419 UP000005203 UP000036403 UP000007267 UP000016666 UP000000311 UP000001593 UP000053825 UP000076502 UP000215335 UP000002358 UP000053605
PRIDE
Pfam
PF01762 Galactosyl_T
ProteinModelPortal
H9JE18
A0A2H1VJ31
A0A2A4K6M7
A0A212EUF8
S4NXU0
A0A437AT78
+ More
A0A437BJX4 A0A194PKX0 A0A0L7LEZ7 A0A194R736 A0A1B6LE51 A0A443QFV8 A0A0P5AJC0 A0A3S4Q954 A0A0P4YW08 A0A0N7ZQZ9 A0A0P5XMH1 A0A0P5J703 A0A0N8D9I0 A0A0P5LSB6 A0A0P5Q469 A0A0P5GUR7 A0A0P5ZZZ2 A0A0P5HLV2 A0A1B6I698 A0A0P5R7P5 A0A1Y3B2A5 E9HTR4 A0A1S3DAY2 A0A0P5RGA7 A0A0P6HVR1 A0A099ZBT2 A0A0P5V9Y5 A0A0P5WXH4 A0A2B4SVP5 A0A067RFU4 A0A1B6EPG0 A0A433TIW4 A0A2I0M2T7 A0A158P1A5 A0A2T7PQF8 A0A195DL48 F4WA33 A0A151I5W5 A0A151IG30 A0A2J7QK23 A0A091HXN7 A0A093GPU1 A0A091NFV3 A0A151XH51 E9IKJ2 A0A1V4KNU9 A0A151K018 U3KJK4 A0A091IM70 G3UU25 A0A091LG01 A0A091F8A8 A0A091RQ55 A0A091KDC8 A0A091JNK1 A0A0T6AWB4 A0A093C9C9 A0A3M0ISV9 A0A091RBA2 H0W6A7 H0Z0U9 A0A091FV83 A0A2P4TG69 A0A250XYI0 A0A091NSA0 A0A226N0L9 A0A091UZY6 A0A0A0AV72 A0A2I0TS58 A0A087VRW8 A0A093IXD0 A0A093H085 A0A093NUM9 A0A1B6D0R3 A0A3L8Q8V4 A0A087QSA9 A0A091US57 A0A093ISC4 A0A226PDQ6 A0A094L8X7 A0A087ZNU1 A0A0J7KDG7 K7EYM3 U3I5J1 E2ATY8 A0A132A8J8 A7SZM8 A0A0L7RAS5 A0A154PA43 A0A094KRT1 A0A232F6A2 A0A091UIZ1 K7JRP2 A0A091WLH4
A0A437BJX4 A0A194PKX0 A0A0L7LEZ7 A0A194R736 A0A1B6LE51 A0A443QFV8 A0A0P5AJC0 A0A3S4Q954 A0A0P4YW08 A0A0N7ZQZ9 A0A0P5XMH1 A0A0P5J703 A0A0N8D9I0 A0A0P5LSB6 A0A0P5Q469 A0A0P5GUR7 A0A0P5ZZZ2 A0A0P5HLV2 A0A1B6I698 A0A0P5R7P5 A0A1Y3B2A5 E9HTR4 A0A1S3DAY2 A0A0P5RGA7 A0A0P6HVR1 A0A099ZBT2 A0A0P5V9Y5 A0A0P5WXH4 A0A2B4SVP5 A0A067RFU4 A0A1B6EPG0 A0A433TIW4 A0A2I0M2T7 A0A158P1A5 A0A2T7PQF8 A0A195DL48 F4WA33 A0A151I5W5 A0A151IG30 A0A2J7QK23 A0A091HXN7 A0A093GPU1 A0A091NFV3 A0A151XH51 E9IKJ2 A0A1V4KNU9 A0A151K018 U3KJK4 A0A091IM70 G3UU25 A0A091LG01 A0A091F8A8 A0A091RQ55 A0A091KDC8 A0A091JNK1 A0A0T6AWB4 A0A093C9C9 A0A3M0ISV9 A0A091RBA2 H0W6A7 H0Z0U9 A0A091FV83 A0A2P4TG69 A0A250XYI0 A0A091NSA0 A0A226N0L9 A0A091UZY6 A0A0A0AV72 A0A2I0TS58 A0A087VRW8 A0A093IXD0 A0A093H085 A0A093NUM9 A0A1B6D0R3 A0A3L8Q8V4 A0A087QSA9 A0A091US57 A0A093ISC4 A0A226PDQ6 A0A094L8X7 A0A087ZNU1 A0A0J7KDG7 K7EYM3 U3I5J1 E2ATY8 A0A132A8J8 A7SZM8 A0A0L7RAS5 A0A154PA43 A0A094KRT1 A0A232F6A2 A0A091UIZ1 K7JRP2 A0A091WLH4
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Golgi apparatus membrane
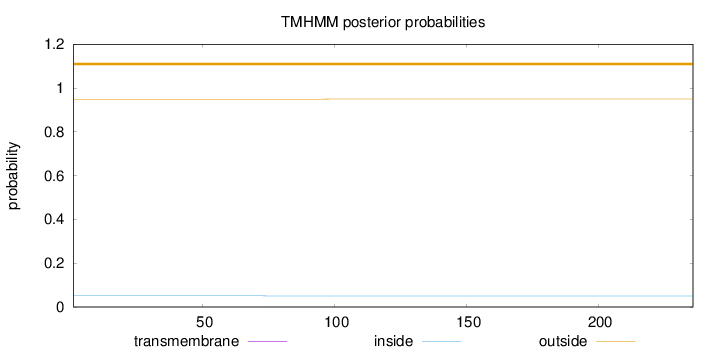
Length:
236
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02314
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05103
outside
1 - 236
Population Genetic Test Statistics
Pi
147.205236
Theta
14.292404
Tajima's D
0.675697
CLR
0
CSRT
0.572771361431928
Interpretation
Uncertain
