Gene
KWMTBOMO09012
Pre Gene Modal
BGIBMGA007774
Annotation
PREDICTED:_decapping_nuclease_DXO_homolog_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.716
Sequence
CDS
ATGCAAATTCAGCCAGAACTTCATGTGAATGCTAGCATTTACTCAAATTCATTTCCTCGATTTGATCGTCCAAAAGTGATAGGATATATTGGTCTCCACAATCTAAAATTTTGTAAAACTTTACGAGACAAAAAAGTGTGTTTTGATTTGAATTTGCATTTAGACAAAGTAAAGCGTAAACCAGTAGATCGGGACGTTAAATTAGATGATTTGTTGCATTTCTTACTTACACAAGAGCACAGATTAAACTTTCCAATACAGAAGACTATTAACGATGCCCCGATTCACTGTTACCGGGGTCTCATGACATGTGTTGCGTGTACACCGTATGAGAATACAGAACCATGGAAAATAGTTACGATTTTACATAAAGGTAACATATACTTATGCGCGAGAGACACTGATGAAAAGAAACATAGAAATCGTAATATGTCTGAGAAAGATAAGCAGTTCACATCATGGGGCTTTAAATTTGAACAATTCATGACATCAGATCATCCTGAAGCCAATCCCAATCCTGATGTACCTGTAGATGAAACAGAAGAATTCTCATTGGTTTTAACTACGAAATTGAACAATCACAAAATAATTTATGGAGCAGAAATGGATGCGATCAGATGTGACAAAGTAGTGGTCCCATCCCCACCATCAAATGATGCTGATCCTCAAACTGTTTTAGAGTATCTGTGTGATAAGGAATTCATTGAGTTAAAAACAAATAGGCATATAGAAAATAATAAACAAGAGATAAATTTCAGGAGGTACAAATGCAAAAAATGGTGGTGTCAATCATTTCTTGTTGGAGTAGACACTATATTATGTGGATTTCGAAATGATAAAGGAATAGTTGAAGAGTTAAAAGTGTTCACAGTAAAAGAATTATCAAAACAAAGTAAGAAATATTGGAGTGGCAATGTTTGCTTTAATTTCTTAGATACATTCTTTACTTACGTAAAAAGATGTCTCATTAGAAGAGTGAAACAAAAATACGGAGAAAAAGCATTGCAAAATTTACATAACTTACCATTAGTTAGTTTACTTTTTGAATGGAATCCTGAAGCACATGTACAAGTTAACGAAGCTTACACATATGAAGATGATCCTATATTACCCGAATTATTTATTAAAAACTATGGTTCTAGAGAACTAATTTGA
Protein
MQIQPELHVNASIYSNSFPRFDRPKVIGYIGLHNLKFCKTLRDKKVCFDLNLHLDKVKRKPVDRDVKLDDLLHFLLTQEHRLNFPIQKTINDAPIHCYRGLMTCVACTPYENTEPWKIVTILHKGNIYLCARDTDEKKHRNRNMSEKDKQFTSWGFKFEQFMTSDHPEANPNPDVPVDETEEFSLVLTTKLNNHKIIYGAEMDAIRCDKVVVPSPPSNDADPQTVLEYLCDKEFIELKTNRHIENNKQEINFRRYKCKKWWCQSFLVGVDTILCGFRNDKGIVEELKVFTVKELSKQSKKYWSGNVCFNFLDTFFTYVKRCLIRRVKQKYGEKALQNLHNLPLVSLLFEWNPEAHVQVNEAYTYEDDPILPELFIKNYGSRELI
Summary
Uniprot
H9JE27
A0A2H1WAP8
A0A2A4K888
A0A3S2LI78
A0A194PJ65
A0A212EQS8
+ More
A0A0L7KJT6 A0A194RCI3 A0A1B6KVL7 A0A067QWW2 A0A1B6JUI1 A0A336L828 A0A0K8TLE2 A0A1B6F1N5 T1JH46 K7IS87 A0A232EKE7 A0A0C9R455 A0A2L2YAR4 A0A2R5L4Z7 Q16FZ8 A0A1Z5L9X8 A0A2P6KPG1 A0A2A4JYK6 A0A1B6CHS4 A0A0N8C8X0 A0A0P5TUU9 A0A0P5B7S5 A0A0N8EA56 A0A0P5UIX0 A0A0P5WYJ8 A0A0P5BGW0 A0A0P5N8R2 A0A0P6CNV9 A0A0A1XCC1 A0A0P5NWW2 A0A2H1WYL6 A0A0N7ZLI1 A0A182FGK5 A0A224YYS4 K1P9Q1 A0A182LV93 A0A0D8XPD7 A0A1X7VSW6 A0A2G8L1B5 A0A210PUF9 A0A182K164 A0A1V9XZA8 A0A0P5MWC2 A0A238BWF2 A0A1D2MCG6 W8BFN3 V3Z2F1 A0A2A4JYW8 A0A1S0U135 A0A182E1D3 A0A1S3K0L4 C3Y9L1
A0A0L7KJT6 A0A194RCI3 A0A1B6KVL7 A0A067QWW2 A0A1B6JUI1 A0A336L828 A0A0K8TLE2 A0A1B6F1N5 T1JH46 K7IS87 A0A232EKE7 A0A0C9R455 A0A2L2YAR4 A0A2R5L4Z7 Q16FZ8 A0A1Z5L9X8 A0A2P6KPG1 A0A2A4JYK6 A0A1B6CHS4 A0A0N8C8X0 A0A0P5TUU9 A0A0P5B7S5 A0A0N8EA56 A0A0P5UIX0 A0A0P5WYJ8 A0A0P5BGW0 A0A0P5N8R2 A0A0P6CNV9 A0A0A1XCC1 A0A0P5NWW2 A0A2H1WYL6 A0A0N7ZLI1 A0A182FGK5 A0A224YYS4 K1P9Q1 A0A182LV93 A0A0D8XPD7 A0A1X7VSW6 A0A2G8L1B5 A0A210PUF9 A0A182K164 A0A1V9XZA8 A0A0P5MWC2 A0A238BWF2 A0A1D2MCG6 W8BFN3 V3Z2F1 A0A2A4JYW8 A0A1S0U135 A0A182E1D3 A0A1S3K0L4 C3Y9L1
Pubmed
EMBL
BABH01002508
ODYU01007404
SOQ50138.1
NWSH01000076
PCG79882.1
RSAL01000109
+ More
RVE47220.1 KQ459602 KPI93367.1 AGBW02013218 OWR43856.1 JTDY01009362 KOB63602.1 KQ460615 KPJ13616.1 GEBQ01024512 JAT15465.1 KK852921 KDR13807.1 GECU01004842 JAT02865.1 UFQS01001980 UFQT01001980 SSX12847.1 SSX32289.1 GDAI01002459 JAI15144.1 GECZ01025607 JAS44162.1 JH432222 NNAY01003801 OXU18836.1 GBYB01011084 JAG80851.1 IAAA01010104 LAA05077.1 GGLE01000425 MBY04551.1 CH478354 EAT33164.1 GFJQ02002747 JAW04223.1 MWRG01007554 PRD28203.1 NWSH01000412 PCG76573.1 GEDC01024375 JAS12923.1 GDIQ01103299 JAL48427.1 GDIP01121688 JAL82026.1 GDIP01189140 JAJ34262.1 GDIQ01046820 JAN47917.1 GDIP01112249 JAL91465.1 GDIP01091849 JAM11866.1 GDIP01199786 JAJ23616.1 GDIQ01145631 JAL06095.1 GDIP01011785 JAM91930.1 GBXI01006184 JAD08108.1 GDIQ01141042 JAL10684.1 ODYU01012016 SOQ58092.1 GDIP01233689 JAI89712.1 GFPF01010939 MAA22085.1 JH818588 EKC20487.1 AXCM01002771 KN716355 KJH46395.1 MRZV01000266 PIK54047.1 NEDP02005488 OWF40092.1 MNPL01001759 OQR78837.1 GDIQ01150230 JAL01496.1 KZ269999 OZC08908.1 LJIJ01001858 ODM90670.1 GAMC01006430 JAC00126.1 KB203412 ESO84783.1 PCG76572.1 JH712118 EFO22715.2 UYRW01000242 VDK64876.1 GG666492 EEN63267.1
RVE47220.1 KQ459602 KPI93367.1 AGBW02013218 OWR43856.1 JTDY01009362 KOB63602.1 KQ460615 KPJ13616.1 GEBQ01024512 JAT15465.1 KK852921 KDR13807.1 GECU01004842 JAT02865.1 UFQS01001980 UFQT01001980 SSX12847.1 SSX32289.1 GDAI01002459 JAI15144.1 GECZ01025607 JAS44162.1 JH432222 NNAY01003801 OXU18836.1 GBYB01011084 JAG80851.1 IAAA01010104 LAA05077.1 GGLE01000425 MBY04551.1 CH478354 EAT33164.1 GFJQ02002747 JAW04223.1 MWRG01007554 PRD28203.1 NWSH01000412 PCG76573.1 GEDC01024375 JAS12923.1 GDIQ01103299 JAL48427.1 GDIP01121688 JAL82026.1 GDIP01189140 JAJ34262.1 GDIQ01046820 JAN47917.1 GDIP01112249 JAL91465.1 GDIP01091849 JAM11866.1 GDIP01199786 JAJ23616.1 GDIQ01145631 JAL06095.1 GDIP01011785 JAM91930.1 GBXI01006184 JAD08108.1 GDIQ01141042 JAL10684.1 ODYU01012016 SOQ58092.1 GDIP01233689 JAI89712.1 GFPF01010939 MAA22085.1 JH818588 EKC20487.1 AXCM01002771 KN716355 KJH46395.1 MRZV01000266 PIK54047.1 NEDP02005488 OWF40092.1 MNPL01001759 OQR78837.1 GDIQ01150230 JAL01496.1 KZ269999 OZC08908.1 LJIJ01001858 ODM90670.1 GAMC01006430 JAC00126.1 KB203412 ESO84783.1 PCG76572.1 JH712118 EFO22715.2 UYRW01000242 VDK64876.1 GG666492 EEN63267.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000037510
+ More
UP000053240 UP000027135 UP000002358 UP000215335 UP000008820 UP000069272 UP000005408 UP000075883 UP000053766 UP000007879 UP000230750 UP000242188 UP000075881 UP000192247 UP000094527 UP000030746 UP000077448 UP000271087 UP000085678 UP000001554
UP000053240 UP000027135 UP000002358 UP000215335 UP000008820 UP000069272 UP000005408 UP000075883 UP000053766 UP000007879 UP000230750 UP000242188 UP000075881 UP000192247 UP000094527 UP000030746 UP000077448 UP000271087 UP000085678 UP000001554
PRIDE
Pfam
PF08652 RAI1
ProteinModelPortal
H9JE27
A0A2H1WAP8
A0A2A4K888
A0A3S2LI78
A0A194PJ65
A0A212EQS8
+ More
A0A0L7KJT6 A0A194RCI3 A0A1B6KVL7 A0A067QWW2 A0A1B6JUI1 A0A336L828 A0A0K8TLE2 A0A1B6F1N5 T1JH46 K7IS87 A0A232EKE7 A0A0C9R455 A0A2L2YAR4 A0A2R5L4Z7 Q16FZ8 A0A1Z5L9X8 A0A2P6KPG1 A0A2A4JYK6 A0A1B6CHS4 A0A0N8C8X0 A0A0P5TUU9 A0A0P5B7S5 A0A0N8EA56 A0A0P5UIX0 A0A0P5WYJ8 A0A0P5BGW0 A0A0P5N8R2 A0A0P6CNV9 A0A0A1XCC1 A0A0P5NWW2 A0A2H1WYL6 A0A0N7ZLI1 A0A182FGK5 A0A224YYS4 K1P9Q1 A0A182LV93 A0A0D8XPD7 A0A1X7VSW6 A0A2G8L1B5 A0A210PUF9 A0A182K164 A0A1V9XZA8 A0A0P5MWC2 A0A238BWF2 A0A1D2MCG6 W8BFN3 V3Z2F1 A0A2A4JYW8 A0A1S0U135 A0A182E1D3 A0A1S3K0L4 C3Y9L1
A0A0L7KJT6 A0A194RCI3 A0A1B6KVL7 A0A067QWW2 A0A1B6JUI1 A0A336L828 A0A0K8TLE2 A0A1B6F1N5 T1JH46 K7IS87 A0A232EKE7 A0A0C9R455 A0A2L2YAR4 A0A2R5L4Z7 Q16FZ8 A0A1Z5L9X8 A0A2P6KPG1 A0A2A4JYK6 A0A1B6CHS4 A0A0N8C8X0 A0A0P5TUU9 A0A0P5B7S5 A0A0N8EA56 A0A0P5UIX0 A0A0P5WYJ8 A0A0P5BGW0 A0A0P5N8R2 A0A0P6CNV9 A0A0A1XCC1 A0A0P5NWW2 A0A2H1WYL6 A0A0N7ZLI1 A0A182FGK5 A0A224YYS4 K1P9Q1 A0A182LV93 A0A0D8XPD7 A0A1X7VSW6 A0A2G8L1B5 A0A210PUF9 A0A182K164 A0A1V9XZA8 A0A0P5MWC2 A0A238BWF2 A0A1D2MCG6 W8BFN3 V3Z2F1 A0A2A4JYW8 A0A1S0U135 A0A182E1D3 A0A1S3K0L4 C3Y9L1
PDB
6AIX
E-value=5.78908e-30,
Score=326
Ontologies
GO
PANTHER
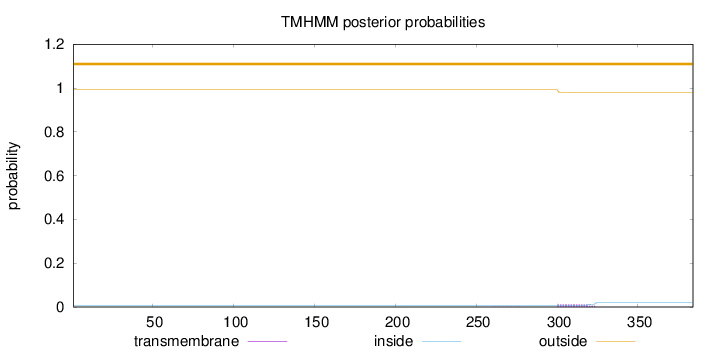
Topology
Length:
384
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.26434
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.00712
outside
1 - 384
Population Genetic Test Statistics
Pi
222.875401
Theta
172.788434
Tajima's D
1.219448
CLR
1.553553
CSRT
0.714164291785411
Interpretation
Uncertain
