Pre Gene Modal
BGIBMGA007776
Annotation
PREDICTED:_exostosin-3_isoform_X1_[Bombyx_mori]
Full name
Exostosin-3
Alternative Name
Protein brother of tout-velu
Location in the cell
PlasmaMembrane Reliability : 3.041
Sequence
CDS
ATGAATATAATATATGAACGTTTTTGTTTGTGGTTGGGACATTTGAAACTATCACGTGTAATAATATTGGTCTCGGTGATATTATTCGTTGTGCCACTTTTTACGCACTACTATTTATCAAAGTATGAATCAATGTCAATATCACAGGGTGACAATGCTTTGAGGTACAATCTCGAGGCATTTAGTGATATATCCACTAAGAATGTTGCGGATCTCAAAGTTAGAATAGAAGAAATGTTAAGAATAAAGGCATCAGTATCAACAGAACTCCGAGAGCTAGAGGCAAAACGAGGTCGCTTACAACGTGAGGCTGCAGCGGCAAGCTCAAAAGCAGACAATGCAAAAGCAGAGTATGTAAGAGCAGCTGCAGAATTACAAAGGCTCAGAGTGTCAGCTGATCAAGCGCGACTGGCCCAGTTAGAAGCAATACGTAGGGATTCACCTGAACTTGCACCACCTATTCAAATAGTGCCGTTACAACCGCCATCAATTCTTCCTTCCTTAAGTCCCAGCAATGAAATAAATTGTAGAATGTTTTCATGTTTTGATCACTCACGATGTTCCCTTACTTCGAGCTTCCCTGTATATCTTTATGATCCCGATATATACACTCCATTAACTGGTGCAGAAGTAGATGGGTTTCTTAAAACTACTTTACGTCAGACTTTAGGATATAATGCTCATCTGACACATAACCCAAATGAGGCCTGTATTTACCTTATTATTGTAGGAGAATCTTTTCCATTTACCAAAGACACTACTACTGTCTCAGACTCCTACAAACTTTATCTCAATGGGTCAGCTATTACTAGCTTACCTTACTGGGGTGGCGATGGGCGTAATCATGTGTTGTTGAATTTAGCCCGCCGTGACTTATCTGTCGGTTCTGGTGATGCTTTTAGAGGAGCATCCACAGGAAGAGCCATGCTAGCTCAATCTACATTTACGCACAATCATTTTAGACCAGGTTTTGATGTAATCACACCACCGGCACTAGGCCCTCCGGGTGGTGATGTTTGGGCAGATTGTGCGCCTATATCTCCCGCAAGAAGAAAGTACTTGCTCAGTTTTCAGGGTTTACAACCACCGCCTAAAGGATTAGACAACGATCAAGATGGAACGTTGATATCATTGCTAGAAAAAATGGTGAAAACAGCACCAGCGTCTGATGTTTTTTTTCTGCAGTTCGAATGTGATCCTCCAGTGGAGAAACGCGTAACTTTGCCGATAGGCGATTGGGCGCTTTGTGGAACGGATAGATCGAGGCGTGCCGTATTGCGTGATTCCACTTTTGTACTCATACTTGCGCCTGCTGATCGGAATTACGTTACAACAGCTCTACTACAGGCTAGATTATATGAAGCGCTCAGATCTGGTGCTATACCTGTCGTCCTAGGCGGTGACAGAATTCGTCTGCCATACGAAGAGGTATTGGATTGGAAACGCGCCGTAATCTTACTGCCGAAAGCGCGGGTCACGGAAATGCATTTTTTAATGAGATCTTTTCCCGATGCCGACCTTTTAGCTCTACGAAGACAGGGTAGAGTACTTTGGGAAAGATATTTGAGTTCAGTTCAGGCCAGCATTGATTCATTGTTGGCAACTATTCGCACCCGCTTACACATTCCTCCAAAACCAGTGCCTCATACCGTCGGGTTGCCGGCTTTTAATGACTCATACTTTCCTCCTCGAGTAGAGCCAGCCGCGGTCGACGCAGAGCCCGAGGAGACGCTAGGTCCGCTGGAGGCTCCGTATTCGAGCCCAACATATCAGCGAAATTTCTCCGTCGGCCTGTTGCACGGATACGAATTATGGAACGAGTGGGGTGAACCTTTCGCTCTGTATCCACAACTACCGTGGGACCCGACGGTGACGTCTGAAGCTCGTTTCTTGGGGTCGGCGGCGGGTTTTCGGCCCGTGGGCGCTGGGGCTGGCGGTTCGGGACGCGAATTTAGCGAAGCCTTGGGTGGAGATCGACCTCGCGAACAATTCACCATAGTCATCCTCACGTATGAACGCGAGGCGGTTTTAGCAGCAGCACTGGCACGATTACGCGGTCTTCCATATCTGAATAAGGTAGTTGTGGTGTGGAATGGTCCTTCGAGCGCAGGAGCGAGTTCTTGGCCGGAGAGCGGCGCGCCAGTGGCGGTGGTGCGTGGCTCGCGGAACTCCCTCAACAACCGTTTCCTGCCTTACCAGCTGATCGAAACCGAGGCCGTACTGTGCGTCGACGACGACGCGCATCTACGACATGACGAGATTATATTCGCGTTCCGCGTCTGGCGCGAGCATCGTGATCAGATTGTGGGCTTTCCCGGTCGTTACCACGCCTGGGATCTTAATTTTAATAATGGATTCCTCTACAATTCGAACTATAGCTGTGAGCTAAGCATGGTGTTGACTGGAGCGGCGTTCGTCCATCGCTACTACTTATGGGCGTACTGGCGAGTACTGCCCGCCGCCATGAGAGATTACGTCGACGAATATATGAACTGCGAAGACATTGCGATGAACTTCCTAGTTGCGCACATCACTAGGAAGCCTCCCGTAAAGGTGACGTCACGATGGACGTTCCGATGTCCCGGTTGTCCGGTTACGTTGTCAGCCGATGAAACACATTTTCACGAACGTCACAAATGTATACAGTTCTTCACACAGGTAATGGGTTACACGCCGCTGTTGTCAACGCAATTCCGAGCTGATTCGGTGCTATTCAAAACGAGGATACCGCACGACAAACAGAAATGTTTCAAGTTCATATAG
Protein
MNIIYERFCLWLGHLKLSRVIILVSVILFVVPLFTHYYLSKYESMSISQGDNALRYNLEAFSDISTKNVADLKVRIEEMLRIKASVSTELRELEAKRGRLQREAAAASSKADNAKAEYVRAAAELQRLRVSADQARLAQLEAIRRDSPELAPPIQIVPLQPPSILPSLSPSNEINCRMFSCFDHSRCSLTSSFPVYLYDPDIYTPLTGAEVDGFLKTTLRQTLGYNAHLTHNPNEACIYLIIVGESFPFTKDTTTVSDSYKLYLNGSAITSLPYWGGDGRNHVLLNLARRDLSVGSGDAFRGASTGRAMLAQSTFTHNHFRPGFDVITPPALGPPGGDVWADCAPISPARRKYLLSFQGLQPPPKGLDNDQDGTLISLLEKMVKTAPASDVFFLQFECDPPVEKRVTLPIGDWALCGTDRSRRAVLRDSTFVLILAPADRNYVTTALLQARLYEALRSGAIPVVLGGDRIRLPYEEVLDWKRAVILLPKARVTEMHFLMRSFPDADLLALRRQGRVLWERYLSSVQASIDSLLATIRTRLHIPPKPVPHTVGLPAFNDSYFPPRVEPAAVDAEPEETLGPLEAPYSSPTYQRNFSVGLLHGYELWNEWGEPFALYPQLPWDPTVTSEARFLGSAAGFRPVGAGAGGSGREFSEALGGDRPREQFTIVILTYEREAVLAAALARLRGLPYLNKVVVVWNGPSSAGASSWPESGAPVAVVRGSRNSLNNRFLPYQLIETEAVLCVDDDAHLRHDEIIFAFRVWREHRDQIVGFPGRYHAWDLNFNNGFLYNSNYSCELSMVLTGAAFVHRYYLWAYWRVLPAAMRDYVDEYMNCEDIAMNFLVAHITRKPPVKVTSRWTFRCPGCPVTLSADETHFHERHKCIQFFTQVMGYTPLLSTQFRADSVLFKTRIPHDKQKCFKFI
Summary
Description
Glycosyltransferase required for the biosynthesis of heparan-sulfate and responsible for the alternating addition of beta-1-4-linked glucuronic acid (GlcA) and alpha-1-4-linked N-acetylglucosamine (GlcNAc) units to nascent heparan sulfate chains. Plays a central role in diffusion of morphogens hedgehog (hh), wingless (wg) and Decapentaplegic (dpp) via its role in heparan sulfate proteoglycans (HSPGs) biosynthesis, HSPGs being required for movement of Hh, Dpp and wg morphogens.
Catalytic Activity
3-O-(beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + UDP-N-acetyl-alpha-D-glucosamine = 3-O-(alpha-D-GlcNAc-(1->4)-beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
Cofactor
Mn(2+)
Subunit
Interacts with sau.
Similarity
Belongs to the glycosyltransferase 47 family.
Keywords
Complete proteome
Developmental protein
Disulfide bond
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
Golgi apparatus
Manganese
Membrane
Metal-binding
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Wnt signaling pathway
Feature
chain Exostosin-3
Uniprot
H9JE29
A0A2A4K6I9
A0A194R7I8
A0A2H1V242
A0A194PKV6
A0A437B9Z5
+ More
A0A212EQT1 A0A1B0DHY7 A0A0L7R4E2 A0A0L7L3E5 A0A1B0CIE1 A0A2A3E3R4 A0A088ARI6 D6WLW9 A0A0C9PY13 A0A154PCP0 K7J5U2 A0A3L8DA57 A0A1Q3FCV2 A0A232EHI4 A0A1Q3FEF2 A0A0T6AW51 A0A0M9A602 A0A151WZA1 A0A195CVJ8 E2C158 A0A1A9YDR4 A0A195BXS6 F4W8W6 A0A158NHG3 A0A1A9UR19 A0A1L8E1C7 A0A195ETX0 A0A1L8E1E3 A0A1L8E1S7 A0A1B0AUV9 A0A182LA04 Q7PY07 A0A182WSQ4 U4UC63 A0A182PSL7 A0A1B0ETL9 A0A182MW04 A0A1I8Q2H4 A0A182UY63 A0A182JZH5 A0A182QDK4 A0A182HLK6 N6U6Q3 A0A182VZJ3 A0A182R8V7 A0A1J1HQB4 A0A182Y3Q2 A0A195DF87 U5EV17 A0A182NRW8 A0A182GUR4 A0A0K8UZR2 A0A1A9WN58 A0A336MS02 A0A084VQK9 A0A0A1WMV9 A0A182JJ75 A0A1I8MLI9 A0A0L0C1K7 Q17P41 A0A182FQP3 A0A026WVW3 W5J584 B4MCN7 A0A0M4EVX8 B4JVM9 A0A1W4V651 B3MCH9 A0A1W4WTE2 Q9XZ08 A0A1B6J4H2 B4PBQ1 A0A182TVJ1 B3NKF2 B4N5Y4 A0A1A9ZCX8 Q28WJ7 A0A0J9RGD6 B4KU72 B4HPB9 A0A0R3NWW8 B0XBE7 A0A1B6CE33 E0VWR0 A0A3R7Q142 A0A0J7NA16 A0A023F4P7 A0A146LNG9 A0A1B6CK88 T1J3I4 X1WRQ0 A0A3B0K7Z3 A0A0U1P5A5 A0A0P5HE35
A0A212EQT1 A0A1B0DHY7 A0A0L7R4E2 A0A0L7L3E5 A0A1B0CIE1 A0A2A3E3R4 A0A088ARI6 D6WLW9 A0A0C9PY13 A0A154PCP0 K7J5U2 A0A3L8DA57 A0A1Q3FCV2 A0A232EHI4 A0A1Q3FEF2 A0A0T6AW51 A0A0M9A602 A0A151WZA1 A0A195CVJ8 E2C158 A0A1A9YDR4 A0A195BXS6 F4W8W6 A0A158NHG3 A0A1A9UR19 A0A1L8E1C7 A0A195ETX0 A0A1L8E1E3 A0A1L8E1S7 A0A1B0AUV9 A0A182LA04 Q7PY07 A0A182WSQ4 U4UC63 A0A182PSL7 A0A1B0ETL9 A0A182MW04 A0A1I8Q2H4 A0A182UY63 A0A182JZH5 A0A182QDK4 A0A182HLK6 N6U6Q3 A0A182VZJ3 A0A182R8V7 A0A1J1HQB4 A0A182Y3Q2 A0A195DF87 U5EV17 A0A182NRW8 A0A182GUR4 A0A0K8UZR2 A0A1A9WN58 A0A336MS02 A0A084VQK9 A0A0A1WMV9 A0A182JJ75 A0A1I8MLI9 A0A0L0C1K7 Q17P41 A0A182FQP3 A0A026WVW3 W5J584 B4MCN7 A0A0M4EVX8 B4JVM9 A0A1W4V651 B3MCH9 A0A1W4WTE2 Q9XZ08 A0A1B6J4H2 B4PBQ1 A0A182TVJ1 B3NKF2 B4N5Y4 A0A1A9ZCX8 Q28WJ7 A0A0J9RGD6 B4KU72 B4HPB9 A0A0R3NWW8 B0XBE7 A0A1B6CE33 E0VWR0 A0A3R7Q142 A0A0J7NA16 A0A023F4P7 A0A146LNG9 A0A1B6CK88 T1J3I4 X1WRQ0 A0A3B0K7Z3 A0A0U1P5A5 A0A0P5HE35
EC Number
2.4.1.223
Pubmed
19121390
26354079
22118469
26227816
18362917
19820115
+ More
20075255 30249741 28648823 20798317 21719571 21347285 20966253 12364791 14747013 17210077 23537049 25244985 26483478 24438588 25830018 25315136 26108605 17510324 24508170 20920257 23761445 17994087 11832488 10731132 12537572 10731138 14645127 14998928 15056609 23720043 17550304 15632085 22936249 20566863 25474469 26823975
20075255 30249741 28648823 20798317 21719571 21347285 20966253 12364791 14747013 17210077 23537049 25244985 26483478 24438588 25830018 25315136 26108605 17510324 24508170 20920257 23761445 17994087 11832488 10731132 12537572 10731138 14645127 14998928 15056609 23720043 17550304 15632085 22936249 20566863 25474469 26823975
EMBL
BABH01002516
BABH01002517
NWSH01000076
PCG79885.1
KQ460615
KPJ13612.1
+ More
ODYU01000314 SOQ34836.1 KQ459602 KPI93364.1 RSAL01000109 RVE47217.1 AGBW02013218 OWR43853.1 AJVK01006080 AJVK01006081 KQ414657 KOC65694.1 JTDY01003287 KOB69799.1 AJWK01013198 KZ288393 PBC26328.1 KQ971343 EFA03391.1 GBYB01006413 JAG76180.1 KQ434870 KZC09591.1 AAZX01005701 QOIP01000011 RLU17214.1 GFDL01009639 JAV25406.1 NNAY01004519 OXU17778.1 GFDL01009115 JAV25930.1 LJIG01022725 KRT79038.1 KQ435727 KOX77940.1 KQ982648 KYQ53224.1 KQ977259 KYN04562.1 GL451853 EFN78322.1 KQ976398 KYM92746.1 GL887973 EGI69358.1 ADTU01001679 GFDF01001556 JAV12528.1 KQ981979 KYN31357.1 GFDF01001618 JAV12466.1 GFDF01001619 JAV12465.1 JXJN01003752 AAAB01008987 EAA00899.4 KB632288 ERL91529.1 AJWK01006508 AXCM01000138 AXCN02000252 APCN01000909 APGK01037417 KB740948 ENN77325.1 CVRI01000009 CRK88662.1 KQ980905 KYN11565.1 GANO01002074 JAB57797.1 JXUM01018889 KQ560525 KXJ81979.1 GDHF01020514 JAI31800.1 UFQT01001064 SSX28718.1 ATLV01015249 KE525004 KFB40253.1 GBXI01014487 JAC99804.1 JRES01001007 KNC26170.1 CH477194 EAT48459.1 KK107078 EZA60195.1 ADMH02002181 ETN58025.1 CH940659 EDW71425.1 CP012524 ALC42125.1 CH916375 EDV98497.1 CH902619 EDV37303.1 AB077850 AE013599 AF132161 GECU01013656 JAS94050.1 CM000158 EDW91535.1 CH954179 EDV55174.1 CH964154 EDW79773.1 CM000071 EAL26670.1 CM002911 KMY94987.1 CH933808 EDW09668.2 CH480816 EDW48557.1 KRT03299.1 DS232632 EDS44259.1 GEDC01025637 GEDC01023240 GEDC01003731 GEDC01002227 JAS11661.1 JAS14058.1 JAS33567.1 JAS35071.1 DS235823 EEB17816.1 QCYY01003030 ROT65837.1 LBMM01007718 KMQ89490.1 GBBI01002749 JAC15963.1 GDHC01018059 GDHC01009171 JAQ00570.1 JAQ09458.1 GEDC01023500 JAS13798.1 JH431827 ABLF02010030 ABLF02010031 OUUW01000947 SPP90194.1 EDY69599.1 GDIQ01251189 JAK00536.1
ODYU01000314 SOQ34836.1 KQ459602 KPI93364.1 RSAL01000109 RVE47217.1 AGBW02013218 OWR43853.1 AJVK01006080 AJVK01006081 KQ414657 KOC65694.1 JTDY01003287 KOB69799.1 AJWK01013198 KZ288393 PBC26328.1 KQ971343 EFA03391.1 GBYB01006413 JAG76180.1 KQ434870 KZC09591.1 AAZX01005701 QOIP01000011 RLU17214.1 GFDL01009639 JAV25406.1 NNAY01004519 OXU17778.1 GFDL01009115 JAV25930.1 LJIG01022725 KRT79038.1 KQ435727 KOX77940.1 KQ982648 KYQ53224.1 KQ977259 KYN04562.1 GL451853 EFN78322.1 KQ976398 KYM92746.1 GL887973 EGI69358.1 ADTU01001679 GFDF01001556 JAV12528.1 KQ981979 KYN31357.1 GFDF01001618 JAV12466.1 GFDF01001619 JAV12465.1 JXJN01003752 AAAB01008987 EAA00899.4 KB632288 ERL91529.1 AJWK01006508 AXCM01000138 AXCN02000252 APCN01000909 APGK01037417 KB740948 ENN77325.1 CVRI01000009 CRK88662.1 KQ980905 KYN11565.1 GANO01002074 JAB57797.1 JXUM01018889 KQ560525 KXJ81979.1 GDHF01020514 JAI31800.1 UFQT01001064 SSX28718.1 ATLV01015249 KE525004 KFB40253.1 GBXI01014487 JAC99804.1 JRES01001007 KNC26170.1 CH477194 EAT48459.1 KK107078 EZA60195.1 ADMH02002181 ETN58025.1 CH940659 EDW71425.1 CP012524 ALC42125.1 CH916375 EDV98497.1 CH902619 EDV37303.1 AB077850 AE013599 AF132161 GECU01013656 JAS94050.1 CM000158 EDW91535.1 CH954179 EDV55174.1 CH964154 EDW79773.1 CM000071 EAL26670.1 CM002911 KMY94987.1 CH933808 EDW09668.2 CH480816 EDW48557.1 KRT03299.1 DS232632 EDS44259.1 GEDC01025637 GEDC01023240 GEDC01003731 GEDC01002227 JAS11661.1 JAS14058.1 JAS33567.1 JAS35071.1 DS235823 EEB17816.1 QCYY01003030 ROT65837.1 LBMM01007718 KMQ89490.1 GBBI01002749 JAC15963.1 GDHC01018059 GDHC01009171 JAQ00570.1 JAQ09458.1 GEDC01023500 JAS13798.1 JH431827 ABLF02010030 ABLF02010031 OUUW01000947 SPP90194.1 EDY69599.1 GDIQ01251189 JAK00536.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000092462 UP000053825 UP000037510 UP000092461 UP000242457 UP000005203 UP000007266 UP000076502 UP000002358 UP000279307 UP000215335 UP000053105 UP000075809 UP000078542 UP000008237 UP000092443 UP000078540 UP000007755 UP000005205 UP000078200 UP000078541 UP000092460 UP000075882 UP000007062 UP000076407 UP000030742 UP000075885 UP000075883 UP000095300 UP000075903 UP000075881 UP000075886 UP000075840 UP000019118 UP000075920 UP000075900 UP000183832 UP000076408 UP000078492 UP000075884 UP000069940 UP000249989 UP000091820 UP000030765 UP000075880 UP000095301 UP000037069 UP000008820 UP000069272 UP000053097 UP000000673 UP000008792 UP000092553 UP000001070 UP000192221 UP000007801 UP000192223 UP000000803 UP000002282 UP000075902 UP000008711 UP000007798 UP000092445 UP000001819 UP000009192 UP000001292 UP000002320 UP000009046 UP000283509 UP000036403 UP000007819 UP000268350
UP000092462 UP000053825 UP000037510 UP000092461 UP000242457 UP000005203 UP000007266 UP000076502 UP000002358 UP000279307 UP000215335 UP000053105 UP000075809 UP000078542 UP000008237 UP000092443 UP000078540 UP000007755 UP000005205 UP000078200 UP000078541 UP000092460 UP000075882 UP000007062 UP000076407 UP000030742 UP000075885 UP000075883 UP000095300 UP000075903 UP000075881 UP000075886 UP000075840 UP000019118 UP000075920 UP000075900 UP000183832 UP000076408 UP000078492 UP000075884 UP000069940 UP000249989 UP000091820 UP000030765 UP000075880 UP000095301 UP000037069 UP000008820 UP000069272 UP000053097 UP000000673 UP000008792 UP000092553 UP000001070 UP000192221 UP000007801 UP000192223 UP000000803 UP000002282 UP000075902 UP000008711 UP000007798 UP000092445 UP000001819 UP000009192 UP000001292 UP000002320 UP000009046 UP000283509 UP000036403 UP000007819 UP000268350
Interpro
SUPFAM
SSF53448
SSF53448
Gene 3D
ProteinModelPortal
H9JE29
A0A2A4K6I9
A0A194R7I8
A0A2H1V242
A0A194PKV6
A0A437B9Z5
+ More
A0A212EQT1 A0A1B0DHY7 A0A0L7R4E2 A0A0L7L3E5 A0A1B0CIE1 A0A2A3E3R4 A0A088ARI6 D6WLW9 A0A0C9PY13 A0A154PCP0 K7J5U2 A0A3L8DA57 A0A1Q3FCV2 A0A232EHI4 A0A1Q3FEF2 A0A0T6AW51 A0A0M9A602 A0A151WZA1 A0A195CVJ8 E2C158 A0A1A9YDR4 A0A195BXS6 F4W8W6 A0A158NHG3 A0A1A9UR19 A0A1L8E1C7 A0A195ETX0 A0A1L8E1E3 A0A1L8E1S7 A0A1B0AUV9 A0A182LA04 Q7PY07 A0A182WSQ4 U4UC63 A0A182PSL7 A0A1B0ETL9 A0A182MW04 A0A1I8Q2H4 A0A182UY63 A0A182JZH5 A0A182QDK4 A0A182HLK6 N6U6Q3 A0A182VZJ3 A0A182R8V7 A0A1J1HQB4 A0A182Y3Q2 A0A195DF87 U5EV17 A0A182NRW8 A0A182GUR4 A0A0K8UZR2 A0A1A9WN58 A0A336MS02 A0A084VQK9 A0A0A1WMV9 A0A182JJ75 A0A1I8MLI9 A0A0L0C1K7 Q17P41 A0A182FQP3 A0A026WVW3 W5J584 B4MCN7 A0A0M4EVX8 B4JVM9 A0A1W4V651 B3MCH9 A0A1W4WTE2 Q9XZ08 A0A1B6J4H2 B4PBQ1 A0A182TVJ1 B3NKF2 B4N5Y4 A0A1A9ZCX8 Q28WJ7 A0A0J9RGD6 B4KU72 B4HPB9 A0A0R3NWW8 B0XBE7 A0A1B6CE33 E0VWR0 A0A3R7Q142 A0A0J7NA16 A0A023F4P7 A0A146LNG9 A0A1B6CK88 T1J3I4 X1WRQ0 A0A3B0K7Z3 A0A0U1P5A5 A0A0P5HE35
A0A212EQT1 A0A1B0DHY7 A0A0L7R4E2 A0A0L7L3E5 A0A1B0CIE1 A0A2A3E3R4 A0A088ARI6 D6WLW9 A0A0C9PY13 A0A154PCP0 K7J5U2 A0A3L8DA57 A0A1Q3FCV2 A0A232EHI4 A0A1Q3FEF2 A0A0T6AW51 A0A0M9A602 A0A151WZA1 A0A195CVJ8 E2C158 A0A1A9YDR4 A0A195BXS6 F4W8W6 A0A158NHG3 A0A1A9UR19 A0A1L8E1C7 A0A195ETX0 A0A1L8E1E3 A0A1L8E1S7 A0A1B0AUV9 A0A182LA04 Q7PY07 A0A182WSQ4 U4UC63 A0A182PSL7 A0A1B0ETL9 A0A182MW04 A0A1I8Q2H4 A0A182UY63 A0A182JZH5 A0A182QDK4 A0A182HLK6 N6U6Q3 A0A182VZJ3 A0A182R8V7 A0A1J1HQB4 A0A182Y3Q2 A0A195DF87 U5EV17 A0A182NRW8 A0A182GUR4 A0A0K8UZR2 A0A1A9WN58 A0A336MS02 A0A084VQK9 A0A0A1WMV9 A0A182JJ75 A0A1I8MLI9 A0A0L0C1K7 Q17P41 A0A182FQP3 A0A026WVW3 W5J584 B4MCN7 A0A0M4EVX8 B4JVM9 A0A1W4V651 B3MCH9 A0A1W4WTE2 Q9XZ08 A0A1B6J4H2 B4PBQ1 A0A182TVJ1 B3NKF2 B4N5Y4 A0A1A9ZCX8 Q28WJ7 A0A0J9RGD6 B4KU72 B4HPB9 A0A0R3NWW8 B0XBE7 A0A1B6CE33 E0VWR0 A0A3R7Q142 A0A0J7NA16 A0A023F4P7 A0A146LNG9 A0A1B6CK88 T1J3I4 X1WRQ0 A0A3B0K7Z3 A0A0U1P5A5 A0A0P5HE35
PDB
1ON8
E-value=1.92462e-23,
Score=274
Ontologies
PATHWAY
GO
PANTHER
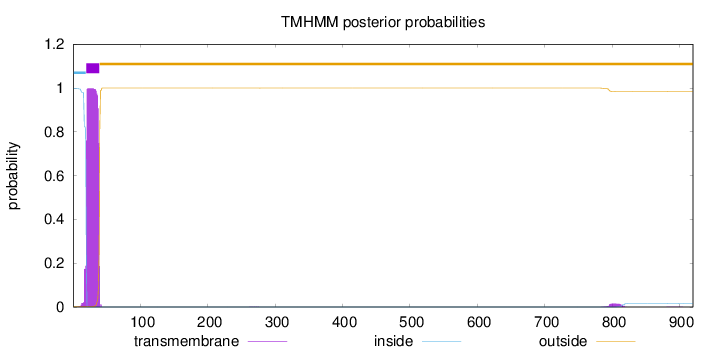
Topology
Subcellular location
Endoplasmic reticulum membrane
Golgi apparatus membrane
Golgi apparatus membrane
Length:
920
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.4878599999999
Exp number, first 60 AAs:
20.15423
Total prob of N-in:
0.99860
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 39
outside
40 - 920
Population Genetic Test Statistics
Pi
329.018876
Theta
200.690719
Tajima's D
1.722537
CLR
0.104403
CSRT
0.834858257087146
Interpretation
Uncertain
