Gene
KWMTBOMO09008 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007757
Annotation
aquaporin_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.426
Sequence
CDS
ATGGGTGAGCTTGGTACTAAACTGGGATTGGACGAGCTGACCGGAGGAGCCGCTACGATCAGCAGAGCACTACTCGCTGAGTTTATCGGTAACTTGCTCCTAAATCTGTTCGGCTGTGGTGCATGTGTGAAGATTTCTCTGGAGTCCAACAGTGAAACGGACATCTTGCTCATCGCTTTGGCGTTTGGGCTAGCGGTATTTGCTGCTGTCTCGGCAATAGGTCATATTTCTGGCGGACACCTAAACCCGGCGGTGACAGCCGGCATGCTGTGCACGGGCCGCATCAAGCTCATCCGTGCGGTGCTGTACGTCATCGTGCAATGCGCCGGGGCCGCGGCCGGTTCGGGACTGCTCAAGGCGCTGACGCCGGACCGCATGGCCGGCAGTCTCGGCTGCACCGGTCTCGGCGTTGACGTTACAGAGCTCCAGGGTTTCGGTATTGAGTTCTTCCTTGGATTCTTACTTGTCTTCATCGTTTGTGGGGTGTGCGATGCCAATAAGCCGGACAGTAAAGCAACGGCCCCGCTCGCCATCGGCCTCACCGTGACCCTGGGTCACTTGCTCGCCGTCGACTACACCGGCTCTGCCATGAACCCAGCCCGCTCCTTTGGCTCGGCCCTCGTCGCTAGCAACTGGAGTCACCATTGGATATACTGGGCTGGACCTATTGCTGGCGGTATTGCTGGTGCTTTGCTGTACGTCCACGGATTTACCGCGCCACCCCAGGAGCCGCCGACCAGATACAGATCTGTTGCCGGTGACGAAAAGGAGCTGAAGCGACTTGATGGTGGTAAACTGGACGACATGGCCTGA
Protein
MGELGTKLGLDELTGGAATISRALLAEFIGNLLLNLFGCGACVKISLESNSETDILLIALAFGLAVFAAVSAIGHISGGHLNPAVTAGMLCTGRIKLIRAVLYVIVQCAGAAAGSGLLKALTPDRMAGSLGCTGLGVDVTELQGFGIEFFLGFLLVFIVCGVCDANKPDSKATAPLAIGLTVTLGHLLAVDYTGSAMNPARSFGSALVASNWSHHWIYWAGPIAGGIAGALLYVHGFTAPPQEPPTRYRSVAGDEKELKRLDGGKLDDMA
Summary
Similarity
Belongs to the MIP/aquaporin (TC 1.A.8) family.
Uniprot
B9ZZR0
H9JE10
A0A2A4K785
A0A2H1V1W6
W8DT73
W8DSL7
+ More
A0A194R8N6 A0A212EQS9 A0A194PK14 T1SQF1 A0A1P8SSF4 A0A023EP10 A0A1Q3FY09 Q1HQY7 A0A2M4BX89 A0A2M4BXC0 T1DQ44 A0A182QW66 W5JRU4 A0A2M3ZH22 A0A2M3Z4Z0 A0A1Q3FEF4 A0A2M4A715 A0A2M4A7E5 A0A182F1Y8 A0A2M4A717 A0A182Y975 A0A182R4A4 A0A1B6CWT4 A0A1Y0AWM5 A0A1L5SAP3 A0A1Q3FXZ5 A0A182QW65 A0A182VU57 A0A182J207 A0A182UKM1 A0A182VAP1 A0A182XDI2 A0A182KNH2 D1MYR4 A0A182I728 A0A182LSW9 A0A182P9Q5 A0A084VYG7 A0A158N7L8 N6U021 A0A182NUN7 V5V0L3 A0A182F1Y9 A0A182R4A3 A0A1Q3FEB2 A0A3Q0IUA2 A0A1L8DEF6 A0A1B6DRL8 A0A1B6CFL2 A0A1W4UIE9 A0A1L8DEH2 A0A1B6ELJ1 A0A182J208 A0A1B0CW61 A0A182VU56 F2YNF6 A0A182VAP0 A0A182KNH1 A0A182XDI3 A0A182I729 A0A182UKM0 A0A182LSX0 A0A1B6J1A5 A0A1B6MCE0 A0A1B6KTE5 A0A182P9Q4 A0A1B6HEZ3 A0A182NUN8 B0X7V5 B4P642 D6WCP1 B4MQJ2 F5HRA2 Q7PWV0 A0A0M4EJ89 F5HRA3 A0A182H914 A0A1I8N5Y2 A0A1W4X1S0 A0A0T6B2J3 A0A182H4C7 G8YY04 A1Z8L8 B4HNI2 B4QBL5 B3NSF7 B3MFC1 B4LQ45 A0A1B6FT45 A0A087ZR22 A0A1Y1M160 A0A1B6M0Y5 A0A1Q3FAH5 E0VTN6
A0A194R8N6 A0A212EQS9 A0A194PK14 T1SQF1 A0A1P8SSF4 A0A023EP10 A0A1Q3FY09 Q1HQY7 A0A2M4BX89 A0A2M4BXC0 T1DQ44 A0A182QW66 W5JRU4 A0A2M3ZH22 A0A2M3Z4Z0 A0A1Q3FEF4 A0A2M4A715 A0A2M4A7E5 A0A182F1Y8 A0A2M4A717 A0A182Y975 A0A182R4A4 A0A1B6CWT4 A0A1Y0AWM5 A0A1L5SAP3 A0A1Q3FXZ5 A0A182QW65 A0A182VU57 A0A182J207 A0A182UKM1 A0A182VAP1 A0A182XDI2 A0A182KNH2 D1MYR4 A0A182I728 A0A182LSW9 A0A182P9Q5 A0A084VYG7 A0A158N7L8 N6U021 A0A182NUN7 V5V0L3 A0A182F1Y9 A0A182R4A3 A0A1Q3FEB2 A0A3Q0IUA2 A0A1L8DEF6 A0A1B6DRL8 A0A1B6CFL2 A0A1W4UIE9 A0A1L8DEH2 A0A1B6ELJ1 A0A182J208 A0A1B0CW61 A0A182VU56 F2YNF6 A0A182VAP0 A0A182KNH1 A0A182XDI3 A0A182I729 A0A182UKM0 A0A182LSX0 A0A1B6J1A5 A0A1B6MCE0 A0A1B6KTE5 A0A182P9Q4 A0A1B6HEZ3 A0A182NUN8 B0X7V5 B4P642 D6WCP1 B4MQJ2 F5HRA2 Q7PWV0 A0A0M4EJ89 F5HRA3 A0A182H914 A0A1I8N5Y2 A0A1W4X1S0 A0A0T6B2J3 A0A182H4C7 G8YY04 A1Z8L8 B4HNI2 B4QBL5 B3NSF7 B3MFC1 B4LQ45 A0A1B6FT45 A0A087ZR22 A0A1Y1M160 A0A1B6M0Y5 A0A1Q3FAH5 E0VTN6
Pubmed
22285686
19121390
26354079
22118469
24945155
17204158
+ More
17510324 20920257 23761445 25244985 28341416 20966253 12364791 24066188 24438588 23537049 21444767 17994087 17550304 18362917 19820115 21497603 26483478 25315136 21993792 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 28004739 20566863
17510324 20920257 23761445 25244985 28341416 20966253 12364791 24066188 24438588 23537049 21444767 17994087 17550304 18362917 19820115 21497603 26483478 25315136 21993792 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 28004739 20566863
EMBL
AB458833
BAH29748.1
BABH01002517
NWSH01000076
PCG79886.1
ODYU01000314
+ More
SOQ34835.1 KC692748 AHF53539.1 KC692749 AHF53540.1 KQ460615 KPJ13610.1 AGBW02013218 OWR43852.1 KQ459602 KPI93363.1 KC999955 AGT50944.1 KX943613 APY26694.1 GAPW01003394 JAC10204.1 GFDL01002600 JAV32445.1 DQ440307 CH477274 ABF18340.1 EAT45186.1 GGFJ01008481 MBW57622.1 GGFJ01008513 MBW57654.1 GAMD01001395 JAB00196.1 AXCN02000734 ADMH02000357 ETN66846.1 GGFM01007072 MBW27823.1 GGFM01002823 MBW23574.1 GFDL01009114 JAV25931.1 GGFK01003211 MBW36532.1 GGFK01003350 MBW36671.1 GGFK01003220 MBW36541.1 GEDC01019391 JAS17907.1 KY921809 ART29398.1 KX129951 APP14132.1 GFDL01002604 JAV32441.1 AAAB01008984 KR020501 AB523397 AKP92849.1 BAI60044.1 APCN01003350 AXCM01000043 ATLV01018366 KE525231 KFB43011.1 APGK01044402 APGK01044403 APGK01044404 APGK01044405 APGK01044406 KB741026 KB631924 ENN74890.1 ERL87183.1 KF649616 AHB86600.1 GFDL01009148 JAV25897.1 GFDF01009242 JAV04842.1 GEDC01028664 GEDC01008986 JAS08634.1 JAS28312.1 GEDC01025097 JAS12201.1 GFDF01009243 JAV04841.1 GECZ01030974 JAS38795.1 AJWK01031823 AJWK01031824 AJWK01031825 JF342682 AEA04450.2 GECU01014742 JAS92964.1 GEBQ01006390 JAT33587.1 GEBQ01025258 JAT14719.1 GECU01034463 JAS73243.1 DS232465 EDS42175.1 CM000158 EDW90917.1 KQ971317 EEZ98804.1 CH963849 EDW74381.1 AB602341 BAK32936.1 EAA14816.4 CP012524 ALC41292.1 AB602342 BAK32937.1 JXUM01030302 KQ560880 KXJ80498.1 LJIG01016112 KRT81580.1 JXUM01001219 FR744897 CBY77924.1 AE013599 AAF58642.1 CH480816 EDW47418.1 CM000362 CM002911 EDX06632.1 KMY93005.1 CH954179 EDV56459.1 CH902619 EDV35595.1 CH940648 EDW61330.1 GECZ01016440 JAS53329.1 GEZM01044913 JAV78045.1 GEBQ01010394 JAT29583.1 GFDL01010552 JAV24493.1 DS235768 EEB16742.1
SOQ34835.1 KC692748 AHF53539.1 KC692749 AHF53540.1 KQ460615 KPJ13610.1 AGBW02013218 OWR43852.1 KQ459602 KPI93363.1 KC999955 AGT50944.1 KX943613 APY26694.1 GAPW01003394 JAC10204.1 GFDL01002600 JAV32445.1 DQ440307 CH477274 ABF18340.1 EAT45186.1 GGFJ01008481 MBW57622.1 GGFJ01008513 MBW57654.1 GAMD01001395 JAB00196.1 AXCN02000734 ADMH02000357 ETN66846.1 GGFM01007072 MBW27823.1 GGFM01002823 MBW23574.1 GFDL01009114 JAV25931.1 GGFK01003211 MBW36532.1 GGFK01003350 MBW36671.1 GGFK01003220 MBW36541.1 GEDC01019391 JAS17907.1 KY921809 ART29398.1 KX129951 APP14132.1 GFDL01002604 JAV32441.1 AAAB01008984 KR020501 AB523397 AKP92849.1 BAI60044.1 APCN01003350 AXCM01000043 ATLV01018366 KE525231 KFB43011.1 APGK01044402 APGK01044403 APGK01044404 APGK01044405 APGK01044406 KB741026 KB631924 ENN74890.1 ERL87183.1 KF649616 AHB86600.1 GFDL01009148 JAV25897.1 GFDF01009242 JAV04842.1 GEDC01028664 GEDC01008986 JAS08634.1 JAS28312.1 GEDC01025097 JAS12201.1 GFDF01009243 JAV04841.1 GECZ01030974 JAS38795.1 AJWK01031823 AJWK01031824 AJWK01031825 JF342682 AEA04450.2 GECU01014742 JAS92964.1 GEBQ01006390 JAT33587.1 GEBQ01025258 JAT14719.1 GECU01034463 JAS73243.1 DS232465 EDS42175.1 CM000158 EDW90917.1 KQ971317 EEZ98804.1 CH963849 EDW74381.1 AB602341 BAK32936.1 EAA14816.4 CP012524 ALC41292.1 AB602342 BAK32937.1 JXUM01030302 KQ560880 KXJ80498.1 LJIG01016112 KRT81580.1 JXUM01001219 FR744897 CBY77924.1 AE013599 AAF58642.1 CH480816 EDW47418.1 CM000362 CM002911 EDX06632.1 KMY93005.1 CH954179 EDV56459.1 CH902619 EDV35595.1 CH940648 EDW61330.1 GECZ01016440 JAS53329.1 GEZM01044913 JAV78045.1 GEBQ01010394 JAT29583.1 GFDL01010552 JAV24493.1 DS235768 EEB16742.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000008820
+ More
UP000075886 UP000000673 UP000069272 UP000076408 UP000075900 UP000075920 UP000075880 UP000075902 UP000075903 UP000076407 UP000075882 UP000075840 UP000075883 UP000075885 UP000030765 UP000019118 UP000030742 UP000075884 UP000079169 UP000192221 UP000092461 UP000002320 UP000002282 UP000007266 UP000007798 UP000007062 UP000092553 UP000069940 UP000249989 UP000095301 UP000192223 UP000000803 UP000001292 UP000000304 UP000008711 UP000007801 UP000008792 UP000005203 UP000009046
UP000075886 UP000000673 UP000069272 UP000076408 UP000075900 UP000075920 UP000075880 UP000075902 UP000075903 UP000076407 UP000075882 UP000075840 UP000075883 UP000075885 UP000030765 UP000019118 UP000030742 UP000075884 UP000079169 UP000192221 UP000092461 UP000002320 UP000002282 UP000007266 UP000007798 UP000007062 UP000092553 UP000069940 UP000249989 UP000095301 UP000192223 UP000000803 UP000001292 UP000000304 UP000008711 UP000007801 UP000008792 UP000005203 UP000009046
Pfam
PF00230 MIP
Interpro
SUPFAM
SSF81338
SSF81338
Gene 3D
CDD
ProteinModelPortal
B9ZZR0
H9JE10
A0A2A4K785
A0A2H1V1W6
W8DT73
W8DSL7
+ More
A0A194R8N6 A0A212EQS9 A0A194PK14 T1SQF1 A0A1P8SSF4 A0A023EP10 A0A1Q3FY09 Q1HQY7 A0A2M4BX89 A0A2M4BXC0 T1DQ44 A0A182QW66 W5JRU4 A0A2M3ZH22 A0A2M3Z4Z0 A0A1Q3FEF4 A0A2M4A715 A0A2M4A7E5 A0A182F1Y8 A0A2M4A717 A0A182Y975 A0A182R4A4 A0A1B6CWT4 A0A1Y0AWM5 A0A1L5SAP3 A0A1Q3FXZ5 A0A182QW65 A0A182VU57 A0A182J207 A0A182UKM1 A0A182VAP1 A0A182XDI2 A0A182KNH2 D1MYR4 A0A182I728 A0A182LSW9 A0A182P9Q5 A0A084VYG7 A0A158N7L8 N6U021 A0A182NUN7 V5V0L3 A0A182F1Y9 A0A182R4A3 A0A1Q3FEB2 A0A3Q0IUA2 A0A1L8DEF6 A0A1B6DRL8 A0A1B6CFL2 A0A1W4UIE9 A0A1L8DEH2 A0A1B6ELJ1 A0A182J208 A0A1B0CW61 A0A182VU56 F2YNF6 A0A182VAP0 A0A182KNH1 A0A182XDI3 A0A182I729 A0A182UKM0 A0A182LSX0 A0A1B6J1A5 A0A1B6MCE0 A0A1B6KTE5 A0A182P9Q4 A0A1B6HEZ3 A0A182NUN8 B0X7V5 B4P642 D6WCP1 B4MQJ2 F5HRA2 Q7PWV0 A0A0M4EJ89 F5HRA3 A0A182H914 A0A1I8N5Y2 A0A1W4X1S0 A0A0T6B2J3 A0A182H4C7 G8YY04 A1Z8L8 B4HNI2 B4QBL5 B3NSF7 B3MFC1 B4LQ45 A0A1B6FT45 A0A087ZR22 A0A1Y1M160 A0A1B6M0Y5 A0A1Q3FAH5 E0VTN6
A0A194R8N6 A0A212EQS9 A0A194PK14 T1SQF1 A0A1P8SSF4 A0A023EP10 A0A1Q3FY09 Q1HQY7 A0A2M4BX89 A0A2M4BXC0 T1DQ44 A0A182QW66 W5JRU4 A0A2M3ZH22 A0A2M3Z4Z0 A0A1Q3FEF4 A0A2M4A715 A0A2M4A7E5 A0A182F1Y8 A0A2M4A717 A0A182Y975 A0A182R4A4 A0A1B6CWT4 A0A1Y0AWM5 A0A1L5SAP3 A0A1Q3FXZ5 A0A182QW65 A0A182VU57 A0A182J207 A0A182UKM1 A0A182VAP1 A0A182XDI2 A0A182KNH2 D1MYR4 A0A182I728 A0A182LSW9 A0A182P9Q5 A0A084VYG7 A0A158N7L8 N6U021 A0A182NUN7 V5V0L3 A0A182F1Y9 A0A182R4A3 A0A1Q3FEB2 A0A3Q0IUA2 A0A1L8DEF6 A0A1B6DRL8 A0A1B6CFL2 A0A1W4UIE9 A0A1L8DEH2 A0A1B6ELJ1 A0A182J208 A0A1B0CW61 A0A182VU56 F2YNF6 A0A182VAP0 A0A182KNH1 A0A182XDI3 A0A182I729 A0A182UKM0 A0A182LSX0 A0A1B6J1A5 A0A1B6MCE0 A0A1B6KTE5 A0A182P9Q4 A0A1B6HEZ3 A0A182NUN8 B0X7V5 B4P642 D6WCP1 B4MQJ2 F5HRA2 Q7PWV0 A0A0M4EJ89 F5HRA3 A0A182H914 A0A1I8N5Y2 A0A1W4X1S0 A0A0T6B2J3 A0A182H4C7 G8YY04 A1Z8L8 B4HNI2 B4QBL5 B3NSF7 B3MFC1 B4LQ45 A0A1B6FT45 A0A087ZR22 A0A1Y1M160 A0A1B6M0Y5 A0A1Q3FAH5 E0VTN6
PDB
2D57
E-value=1.97763e-30,
Score=328
Ontologies
GO
PANTHER
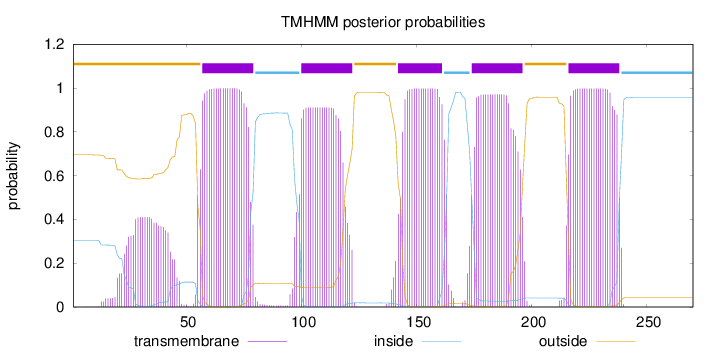
Topology
Length:
270
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
115.02255
Exp number, first 60 AAs:
14.10113
Total prob of N-in:
0.30517
POSSIBLE N-term signal
sequence
outside
1 - 56
TMhelix
57 - 79
inside
80 - 99
TMhelix
100 - 122
outside
123 - 141
TMhelix
142 - 161
inside
162 - 173
TMhelix
174 - 196
outside
197 - 215
TMhelix
216 - 238
inside
239 - 270
Population Genetic Test Statistics
Pi
267.812377
Theta
172.553982
Tajima's D
1.729268
CLR
0.418988
CSRT
0.836558172091395
Interpretation
Uncertain
