Gene
KWMTBOMO09004
Annotation
PREDICTED:_multiple_C2_and_transmembrane_domain-containing_protein_1-like_isoform_X1_[Bombyx_mori]
Full name
Multiple C2 and transmembrane domain-containing protein
Location in the cell
PlasmaMembrane Reliability : 2.536
Sequence
CDS
ATGTATCCGATCCGGTACTACAATGAAGTGTCGTTCCCGATCCCGGACGGTTTCGGTATGGATCCGATGGTACCGAACACGCTGGTGGTATCAGCAGACATCATGGACCGGATCGGAAGCGGACTTTCGTTGCTCCGAGATCACGGAAAAAAAGTACAAAAGTACGTGTGGAAAAACAGAAAATTTGACGTACTGAAGAAGCCTACGAAATCTTTAGTAAATATCGTATTAATAGAAGCTAAAGACTTGCCGGATGACCAGACTACAACCTGCAATGGCCTCTACTGCAAGTTCAAGTTAGGCAACGAATCTCACAAATCTAGACAAGTATTGAAAACAAAGCCTGCATGGTGTGAGAGATTCAATATATATTTGTACGAGGAAAACAATTTAGAGGTATCCTTGTGGCATAAAGGAAAACAGAAAAACTTTATGGGAAGATGCGTCATTGACCTTTCTCGACTCGAAAAAGAAAAGACTCACGATATTTGGCAGGAATTGGAATGTGGATACGGTTTTATACATATGTTGATCACGGTCAACGGATCACAGTTAACCAGAGGATTGGCTGTTGACAATATACCGACAAGCAACAACGTTTTACACCCAACACCAAATATTGAAGAATTTTCATGGTATCGCCTAAAGAACAATTGGAATGAAGTGGGTCAGTTGTCAGTGACTGTATACGGCGCCAGAGGGCTAAGTCCACTCGGGATCAGCGGAAAGGCTGATGCATATTGCGTTTTAGTCTTGGATAACTCAAGGATACAGACTCACACAGTCCGTGGCGCGGCAGAGCCGATGTGGAACAAAAGTTATACATTTGCCGTAAACGATATTACGTCAACCCTGGATATCAAAGTTCACGATGAATCGATTATCGGTGAAACTCTCGGTAAAATATCGATACCACTCTTACGAATCATCAACGAAGAGAAAAGATGGTACGCTCTAAAAGATAAAACGAAAAAGTCGAATGCGAAGGGAAACTGCCCCAGGATACTCCTGCAGATGTCCATTGTATGGAATCCGTTAAAAGCTGCGCTACGAACTTTAAGTCCTAAGGAAACGAAATACGATCAGAAGACGCCTAAAAAATTTAGTATTCCCTTAATATATAACAATCTGCTATTCATCAAAGATATATTTAATGTGGTACATACAGGGAACGAATATTTAAAGAATTCATTTGAATGGGAGAACAGAGAGAAGTCGGTGCTAGCGTTATGCGCTTGGCTGGTCTTCTGGTTCTTTTTCCGAATGTGGATGACACCGCTGCTAGTTCTTCTGCCGTTCCTTTACTACTGGACCGGTCACCGCAGCAATAGTACTACTAATTCATTAAGTAACATTTACGGATCCGAGGATGAATTATCCGAAGAGGATCTTGAATCAACGAAGGATGAAAAAACGATTAAAACCCGTCTCTACGGATTACAGGAATTGACGTTTACAATAAAAGACGGCATCGATTACATAGTGTCACTTTTAGAGCGAGTTAAAAATTTATTGAATTTCACTATCCCGTACCTAAGTTATTTAGCGATGGCCGCAATGGTGGGTCTCAGCGTGGCGTTTTATCTAATACCTGCGAACTACATCTTCATGGCATTGGGTATTTATAAATTTACTCGTAAATACTTGAATCCCGATCGAGTTCCGAACAATGATATCTTGGATTTCATATCGAGGGTGCCCGACGATAGGACACTGAAACAATGGCAGGAATTGAAAGTACCCGAACCGAGCTTAAGCAGGACAGGATCAAAGTGCAGAAGATAA
Protein
MYPIRYYNEVSFPIPDGFGMDPMVPNTLVVSADIMDRIGSGLSLLRDHGKKVQKYVWKNRKFDVLKKPTKSLVNIVLIEAKDLPDDQTTTCNGLYCKFKLGNESHKSRQVLKTKPAWCERFNIYLYEENNLEVSLWHKGKQKNFMGRCVIDLSRLEKEKTHDIWQELECGYGFIHMLITVNGSQLTRGLAVDNIPTSNNVLHPTPNIEEFSWYRLKNNWNEVGQLSVTVYGARGLSPLGISGKADAYCVLVLDNSRIQTHTVRGAAEPMWNKSYTFAVNDITSTLDIKVHDESIIGETLGKISIPLLRIINEEKRWYALKDKTKKSNAKGNCPRILLQMSIVWNPLKAALRTLSPKETKYDQKTPKKFSIPLIYNNLLFIKDIFNVVHTGNEYLKNSFEWENREKSVLALCAWLVFWFFFRMWMTPLLVLLPFLYYWTGHRSNSTTNSLSNIYGSEDELSEEDLESTKDEKTIKTRLYGLQELTFTIKDGIDYIVSLLERVKNLLNFTIPYLSYLAMAAMVGLSVAFYLIPANYIFMALGIYKFTRKYLNPDRVPNNDILDFISRVPDDRTLKQWQELKVPEPSLSRTGSKCRR
Summary
Description
Calcium sensor which is essential for the stabilization of normal baseline neurotransmitter release and for the induction and long-term maintenance of presynaptic homeostatic plasticity (PubMed:28485711).
Cofactor
Ca(2+)
Keywords
Alternative splicing
Calcium
Complete proteome
Endoplasmic reticulum
Membrane
Reference proteome
Repeat
Transmembrane
Transmembrane helix
Feature
chain Multiple C2 and transmembrane domain-containing protein
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4J601
A0A2H1V383
A0A2A4J4U8
A0A194R7I2
A0A3S2NMQ7
A0A194PQF0
+ More
A0A212F6Q9 A0A2A4J5L1 A0A1E1W180 A0A3S2L1S9 A0A2A4J510 A0A2A4K8X0 A0A194R714 A0A2H1WBS6 H9JW63 A0A2A4K984 A0A2W1BKK6 A0A437BN69 A0A067QZE1 E0VQX2 A0A1Y1M3S5 A0A2J7QM48 A0A1Y1M0D1 U4UKD4 T1I1Y3 A0A0N0P9T6 N6TJP3 A0A232FFH3 A0A1Y1M5V4 Q174I5 A0A1E1X4F5 A0A1I8MDV2 A0A1E1XKP4 A0A1I8PKK1 A0A182GQ57 A0A0P6IW11 A0A1Q3G4S2 A0A1Q3G4S1 A0A1I8MDU5 A0A1S4FEV1 A0A1Q3G4R3 A0A1Q3G4Q9 A0A1E1WBJ9 A0A1I8PKP5 A0A0K8VPT3 A0A034VM56 A0A336MFH5 W8B6U0 A0A3S2M9K5 A0A182X6G0 A0A084VFA7 A0A182UEL5 A0A182VKM5 A0A182I3R3 A0A1S4GVR1 A0A182KRV2 A0A182JQQ1 A0A182RFQ2 A0A182P9V6 A0A0A1XHY1 A0A182J121 A0A182WIT4 A0A182XZH8 A0A182MA45 A0A182Q4S6 A0A182NHJ8 B4QD30 Q7JZN2 E9GTE2 B4HP15 A0A210PMT3 A0A1A9XHA5 B4GA37 Q5TX35 A1ZBD6-3 A1ZBD6 A1ZBD6-2 A0A1B6CV73 A0A341BLU7 A0A0P5PPJ2 B3MI22 A0A341BIY8 A0A0J9RG79 A0A3Q1EAQ1 B3NKB2 A0A341BIZ4 A0A340YBK5 A0A2U4AW94 A0A341BIZ3 A0A341BKF4 A0A1A9WYX3 A0A2U4AWR9 A0A2U4AWA2 A0A384BDH7 A0A341BLU3 A0A384BDZ8 A0A384BCQ7 A0A384BCV7
A0A212F6Q9 A0A2A4J5L1 A0A1E1W180 A0A3S2L1S9 A0A2A4J510 A0A2A4K8X0 A0A194R714 A0A2H1WBS6 H9JW63 A0A2A4K984 A0A2W1BKK6 A0A437BN69 A0A067QZE1 E0VQX2 A0A1Y1M3S5 A0A2J7QM48 A0A1Y1M0D1 U4UKD4 T1I1Y3 A0A0N0P9T6 N6TJP3 A0A232FFH3 A0A1Y1M5V4 Q174I5 A0A1E1X4F5 A0A1I8MDV2 A0A1E1XKP4 A0A1I8PKK1 A0A182GQ57 A0A0P6IW11 A0A1Q3G4S2 A0A1Q3G4S1 A0A1I8MDU5 A0A1S4FEV1 A0A1Q3G4R3 A0A1Q3G4Q9 A0A1E1WBJ9 A0A1I8PKP5 A0A0K8VPT3 A0A034VM56 A0A336MFH5 W8B6U0 A0A3S2M9K5 A0A182X6G0 A0A084VFA7 A0A182UEL5 A0A182VKM5 A0A182I3R3 A0A1S4GVR1 A0A182KRV2 A0A182JQQ1 A0A182RFQ2 A0A182P9V6 A0A0A1XHY1 A0A182J121 A0A182WIT4 A0A182XZH8 A0A182MA45 A0A182Q4S6 A0A182NHJ8 B4QD30 Q7JZN2 E9GTE2 B4HP15 A0A210PMT3 A0A1A9XHA5 B4GA37 Q5TX35 A1ZBD6-3 A1ZBD6 A1ZBD6-2 A0A1B6CV73 A0A341BLU7 A0A0P5PPJ2 B3MI22 A0A341BIY8 A0A0J9RG79 A0A3Q1EAQ1 B3NKB2 A0A341BIZ4 A0A340YBK5 A0A2U4AW94 A0A341BIZ3 A0A341BKF4 A0A1A9WYX3 A0A2U4AWR9 A0A2U4AWA2 A0A384BDH7 A0A341BLU3 A0A384BDZ8 A0A384BCQ7 A0A384BCV7
Pubmed
26354079
22118469
19121390
28756777
24845553
20566863
+ More
28004739 23537049 28648823 17510324 28503490 25315136 29209593 26483478 26999592 25348373 24495485 24438588 12364791 20966253 25830018 25244985 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 28812685 28485711 22936249
28004739 23537049 28648823 17510324 28503490 25315136 29209593 26483478 26999592 25348373 24495485 24438588 12364791 20966253 25830018 25244985 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 28812685 28485711 22936249
EMBL
NWSH01002972
PCG67166.1
ODYU01000314
SOQ34832.1
PCG67167.1
KQ460615
+ More
KPJ13607.1 RSAL01000237 RVE43708.1 KQ459602 KPI93360.1 AGBW02009983 OWR49417.1 PCG67169.1 GDQN01010306 JAT80748.1 RVE43709.1 PCG67165.1 NWSH01000040 PCG80354.1 KPJ13608.1 ODYU01007596 SOQ50513.1 BABH01025337 BABH01025338 PCG80353.1 KZ150109 PZC73376.1 RSAL01000029 RVE51755.1 KK852859 KDR14857.1 DS235442 EEB15778.1 GEZM01043132 JAV79290.1 NEVH01013215 PNF29689.1 GEZM01043131 JAV79292.1 KB632339 ERL92928.1 ACPB03005944 KQ459166 KPJ03575.1 APGK01034847 APGK01034848 APGK01034849 APGK01034850 KB740923 ENN78073.1 NNAY01000316 OXU29270.1 GEZM01043135 JAV79286.1 CH477410 EAT41473.1 GFAC01005038 JAT94150.1 GFAA01003656 JAT99778.1 JXUM01079729 JXUM01079730 JXUM01079731 JXUM01079732 GDUN01000054 JAN95865.1 GFDL01000252 JAV34793.1 GFDL01000251 JAV34794.1 GFDL01000254 JAV34791.1 GFDL01000258 JAV34787.1 GDQN01006737 JAT84317.1 GDHF01011734 JAI40580.1 GAKP01015775 JAC43177.1 UFQT01001061 SSX28680.1 GAMC01021074 GAMC01021071 JAB85484.1 RSAL01000007 RVE54120.1 ATLV01012371 KE524785 KFB36651.1 APCN01000218 AAAB01008807 GBXI01009905 GBXI01009819 GBXI01003706 JAD04387.1 JAD04473.1 JAD10586.1 AXCM01001910 AXCN02000592 CM000362 EDX07735.1 AE013599 AY071153 AAF57640.2 AAL48775.1 GL732563 EFX77264.1 CH480816 EDW48517.1 NEDP02005580 OWF37787.1 CH479181 EDW31789.1 EAL41731.2 GEDC01020165 GEDC01015264 JAS17133.1 JAS22034.1 GDIQ01131074 JAL20652.1 CH902619 EDV38032.2 CM002911 KMY94937.1 CH954179 EDV55134.2
KPJ13607.1 RSAL01000237 RVE43708.1 KQ459602 KPI93360.1 AGBW02009983 OWR49417.1 PCG67169.1 GDQN01010306 JAT80748.1 RVE43709.1 PCG67165.1 NWSH01000040 PCG80354.1 KPJ13608.1 ODYU01007596 SOQ50513.1 BABH01025337 BABH01025338 PCG80353.1 KZ150109 PZC73376.1 RSAL01000029 RVE51755.1 KK852859 KDR14857.1 DS235442 EEB15778.1 GEZM01043132 JAV79290.1 NEVH01013215 PNF29689.1 GEZM01043131 JAV79292.1 KB632339 ERL92928.1 ACPB03005944 KQ459166 KPJ03575.1 APGK01034847 APGK01034848 APGK01034849 APGK01034850 KB740923 ENN78073.1 NNAY01000316 OXU29270.1 GEZM01043135 JAV79286.1 CH477410 EAT41473.1 GFAC01005038 JAT94150.1 GFAA01003656 JAT99778.1 JXUM01079729 JXUM01079730 JXUM01079731 JXUM01079732 GDUN01000054 JAN95865.1 GFDL01000252 JAV34793.1 GFDL01000251 JAV34794.1 GFDL01000254 JAV34791.1 GFDL01000258 JAV34787.1 GDQN01006737 JAT84317.1 GDHF01011734 JAI40580.1 GAKP01015775 JAC43177.1 UFQT01001061 SSX28680.1 GAMC01021074 GAMC01021071 JAB85484.1 RSAL01000007 RVE54120.1 ATLV01012371 KE524785 KFB36651.1 APCN01000218 AAAB01008807 GBXI01009905 GBXI01009819 GBXI01003706 JAD04387.1 JAD04473.1 JAD10586.1 AXCM01001910 AXCN02000592 CM000362 EDX07735.1 AE013599 AY071153 AAF57640.2 AAL48775.1 GL732563 EFX77264.1 CH480816 EDW48517.1 NEDP02005580 OWF37787.1 CH479181 EDW31789.1 EAL41731.2 GEDC01020165 GEDC01015264 JAS17133.1 JAS22034.1 GDIQ01131074 JAL20652.1 CH902619 EDV38032.2 CM002911 KMY94937.1 CH954179 EDV55134.2
Proteomes
UP000218220
UP000053240
UP000283053
UP000053268
UP000007151
UP000005204
+ More
UP000027135 UP000009046 UP000235965 UP000030742 UP000015103 UP000019118 UP000215335 UP000008820 UP000095301 UP000095300 UP000069940 UP000076407 UP000030765 UP000075902 UP000075903 UP000075840 UP000075882 UP000075881 UP000075900 UP000075885 UP000075880 UP000075920 UP000076408 UP000075883 UP000075886 UP000075884 UP000000304 UP000000803 UP000000305 UP000001292 UP000242188 UP000092443 UP000008744 UP000007062 UP000252040 UP000007801 UP000257200 UP000008711 UP000265300 UP000245320 UP000091820 UP000261681
UP000027135 UP000009046 UP000235965 UP000030742 UP000015103 UP000019118 UP000215335 UP000008820 UP000095301 UP000095300 UP000069940 UP000076407 UP000030765 UP000075902 UP000075903 UP000075840 UP000075882 UP000075881 UP000075900 UP000075885 UP000075880 UP000075920 UP000076408 UP000075883 UP000075886 UP000075884 UP000000304 UP000000803 UP000000305 UP000001292 UP000242188 UP000092443 UP000008744 UP000007062 UP000252040 UP000007801 UP000257200 UP000008711 UP000265300 UP000245320 UP000091820 UP000261681
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J601
A0A2H1V383
A0A2A4J4U8
A0A194R7I2
A0A3S2NMQ7
A0A194PQF0
+ More
A0A212F6Q9 A0A2A4J5L1 A0A1E1W180 A0A3S2L1S9 A0A2A4J510 A0A2A4K8X0 A0A194R714 A0A2H1WBS6 H9JW63 A0A2A4K984 A0A2W1BKK6 A0A437BN69 A0A067QZE1 E0VQX2 A0A1Y1M3S5 A0A2J7QM48 A0A1Y1M0D1 U4UKD4 T1I1Y3 A0A0N0P9T6 N6TJP3 A0A232FFH3 A0A1Y1M5V4 Q174I5 A0A1E1X4F5 A0A1I8MDV2 A0A1E1XKP4 A0A1I8PKK1 A0A182GQ57 A0A0P6IW11 A0A1Q3G4S2 A0A1Q3G4S1 A0A1I8MDU5 A0A1S4FEV1 A0A1Q3G4R3 A0A1Q3G4Q9 A0A1E1WBJ9 A0A1I8PKP5 A0A0K8VPT3 A0A034VM56 A0A336MFH5 W8B6U0 A0A3S2M9K5 A0A182X6G0 A0A084VFA7 A0A182UEL5 A0A182VKM5 A0A182I3R3 A0A1S4GVR1 A0A182KRV2 A0A182JQQ1 A0A182RFQ2 A0A182P9V6 A0A0A1XHY1 A0A182J121 A0A182WIT4 A0A182XZH8 A0A182MA45 A0A182Q4S6 A0A182NHJ8 B4QD30 Q7JZN2 E9GTE2 B4HP15 A0A210PMT3 A0A1A9XHA5 B4GA37 Q5TX35 A1ZBD6-3 A1ZBD6 A1ZBD6-2 A0A1B6CV73 A0A341BLU7 A0A0P5PPJ2 B3MI22 A0A341BIY8 A0A0J9RG79 A0A3Q1EAQ1 B3NKB2 A0A341BIZ4 A0A340YBK5 A0A2U4AW94 A0A341BIZ3 A0A341BKF4 A0A1A9WYX3 A0A2U4AWR9 A0A2U4AWA2 A0A384BDH7 A0A341BLU3 A0A384BDZ8 A0A384BCQ7 A0A384BCV7
A0A212F6Q9 A0A2A4J5L1 A0A1E1W180 A0A3S2L1S9 A0A2A4J510 A0A2A4K8X0 A0A194R714 A0A2H1WBS6 H9JW63 A0A2A4K984 A0A2W1BKK6 A0A437BN69 A0A067QZE1 E0VQX2 A0A1Y1M3S5 A0A2J7QM48 A0A1Y1M0D1 U4UKD4 T1I1Y3 A0A0N0P9T6 N6TJP3 A0A232FFH3 A0A1Y1M5V4 Q174I5 A0A1E1X4F5 A0A1I8MDV2 A0A1E1XKP4 A0A1I8PKK1 A0A182GQ57 A0A0P6IW11 A0A1Q3G4S2 A0A1Q3G4S1 A0A1I8MDU5 A0A1S4FEV1 A0A1Q3G4R3 A0A1Q3G4Q9 A0A1E1WBJ9 A0A1I8PKP5 A0A0K8VPT3 A0A034VM56 A0A336MFH5 W8B6U0 A0A3S2M9K5 A0A182X6G0 A0A084VFA7 A0A182UEL5 A0A182VKM5 A0A182I3R3 A0A1S4GVR1 A0A182KRV2 A0A182JQQ1 A0A182RFQ2 A0A182P9V6 A0A0A1XHY1 A0A182J121 A0A182WIT4 A0A182XZH8 A0A182MA45 A0A182Q4S6 A0A182NHJ8 B4QD30 Q7JZN2 E9GTE2 B4HP15 A0A210PMT3 A0A1A9XHA5 B4GA37 Q5TX35 A1ZBD6-3 A1ZBD6 A1ZBD6-2 A0A1B6CV73 A0A341BLU7 A0A0P5PPJ2 B3MI22 A0A341BIY8 A0A0J9RG79 A0A3Q1EAQ1 B3NKB2 A0A341BIZ4 A0A340YBK5 A0A2U4AW94 A0A341BIZ3 A0A341BKF4 A0A1A9WYX3 A0A2U4AWR9 A0A2U4AWA2 A0A384BDH7 A0A341BLU3 A0A384BDZ8 A0A384BCQ7 A0A384BCV7
PDB
2EP6
E-value=8.87462e-17,
Score=214
Ontologies
KEGG
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
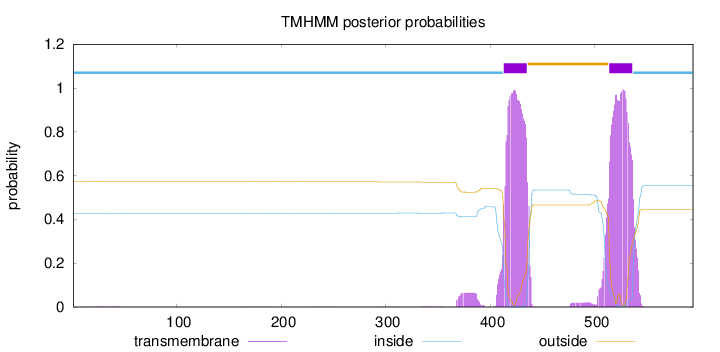
Length:
594
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
49.10471
Exp number, first 60 AAs:
0.04614
Total prob of N-in:
0.42549
inside
1 - 412
TMhelix
413 - 435
outside
436 - 513
TMhelix
514 - 536
inside
537 - 594
Population Genetic Test Statistics
Pi
218.813998
Theta
172.153823
Tajima's D
1.121513
CLR
0
CSRT
0.690415479226039
Interpretation
Uncertain
