Pre Gene Modal
BGIBMGA007778
Annotation
PREDICTED:_golgi_SNAP_receptor_complex_member_2_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.476
Sequence
CDS
ATGGAAACATTATACCATCAGACAAATCAATTGCTACAAGAAACTACAGATCTTTTTAATAAATTAGAGAGAGATCCAGCCAATCACCAACAAATAGAAAATGCAATTCAATCAAAGATAAACACAGTCAATGCGAACTGTGAAAAACTAGATATTTATGTTTTTAAAACACCAATAAATCAAAGGTCTATGGCTAAAATGAGAGTGGACCAGCTGAAGTATGATAACAAACACATACAGGCTTCTCTTATAAATGCCCAAAACAAAAGACAAAAACGGGAACAAGAATTAGCTGAAAGGGAACAACTTTTGAACCGGCGTTTTGGACATGATCACACAGAAATAAATGTAGACTACTTAGCTCAAGAGCAAGCGTCTTTGCAGAATTCACATAGACATGTCGATGATATGTTGCATACAGGTACGAATATTTTGAACACGCTTAAATATAACAGAGACACGATCAAGAGTGCCCATAGGAGGATCATCGATTTGGCCAATACCCTGGGCTTGTCTAATGCGACTATCTCACTGATAGAACGCAGAGTTTCCCAGGACAAATATATTTTGTTTGGCGGAATGATATTCACTTTGGGTGTTATCGTTCTTGTTGTTATATACTTGACGTAG
Protein
METLYHQTNQLLQETTDLFNKLERDPANHQQIENAIQSKINTVNANCEKLDIYVFKTPINQRSMAKMRVDQLKYDNKHIQASLINAQNKRQKREQELAEREQLLNRRFGHDHTEINVDYLAQEQASLQNSHRHVDDMLHTGTNILNTLKYNRDTIKSAHRRIIDLANTLGLSNATISLIERRVSQDKYILFGGMIFTLGVIVLVVIYLT
Summary
Description
Involved in transport of proteins from the cis/medial-Golgi to the trans-Golgi network.
Similarity
Belongs to the GOSR2 family.
Uniprot
H9JE31
Q1HQ00
A0A2A4J5B7
A0A1E1WMJ1
A0A194PK09
A0A212F6Q2
+ More
I4DPT7 A0A194R8N1 S4PMX3 A0A3S2LTP6 D6WM18 A0A0K8TS74 A0A026WZ20 A0A1W4X3S9 A0A088AM34 A0A154P2L7 A0A1Y1LR73 A0A2A3EEV0 A0A0N0BIR7 A0A1L8DUP4 A0A1B6DPV0 Q17AQ3 A0A1S4F9W4 A0A1Q3FC92 A0A023ELX1 A0A2J7QDZ7 A0A182J0I4 A0A084VJM4 B0WGZ5 A0A182M6V1 E9J3D6 A0A182XXE0 A0A1B0D9D3 A0A182I8T3 A0A182P786 A0A182UVX7 A0A182R5X2 A0A182WV58 A0A1S4GJ96 A0A182LAP9 A0A182UA30 A0A1Y9H2L8 A0A1B6IEY5 A0A2M4AVR4 A0A1I8JSN3 A0A195B6K8 A0A158NGY9 A0A195FNG0 A0A1B6EQ79 A0A151K2Z8 A0A151XD97 A0A2M4C032 A0A1B6IA95 A0A0L7RIF6 F4X136 A0A1B6LIN1 A0A087YXN2 A0A2M4CQE7 A0A182K4T4 A0A182Q7A6 A0A182VXV1 A0A2P8Y8U6 A0A2M4CYI4 J3JZ30 A0A151I8K0 A0A1L8DW11 A0A0P4VL02 A0A0A9WY87 E2BW95 A0A0A9YXC4 U5EZ35 E2ALZ0 A0A0T6AYV2 T1HUY8 A0A0V0G7A0 A0A0K8U5V0 A0A1I8PZ29 A0A1B0GGE7 A0A1A9ZMI1 A0A224YU72 A0A131YCB8 A0A034WGR1 A0A210QMS3 A0A1I8MRY9 W8BU74 A0A023FFN8 A0A1A9WR99 A0A1S3J6A9 A0A087TI67 K7IWE6 A0A232FD88 B4MM12 B4KXE5 A0A0K8RMG3 A0A0L8GRT4 B4IX16 R7TL43 A0A336LPP1 V4B413
I4DPT7 A0A194R8N1 S4PMX3 A0A3S2LTP6 D6WM18 A0A0K8TS74 A0A026WZ20 A0A1W4X3S9 A0A088AM34 A0A154P2L7 A0A1Y1LR73 A0A2A3EEV0 A0A0N0BIR7 A0A1L8DUP4 A0A1B6DPV0 Q17AQ3 A0A1S4F9W4 A0A1Q3FC92 A0A023ELX1 A0A2J7QDZ7 A0A182J0I4 A0A084VJM4 B0WGZ5 A0A182M6V1 E9J3D6 A0A182XXE0 A0A1B0D9D3 A0A182I8T3 A0A182P786 A0A182UVX7 A0A182R5X2 A0A182WV58 A0A1S4GJ96 A0A182LAP9 A0A182UA30 A0A1Y9H2L8 A0A1B6IEY5 A0A2M4AVR4 A0A1I8JSN3 A0A195B6K8 A0A158NGY9 A0A195FNG0 A0A1B6EQ79 A0A151K2Z8 A0A151XD97 A0A2M4C032 A0A1B6IA95 A0A0L7RIF6 F4X136 A0A1B6LIN1 A0A087YXN2 A0A2M4CQE7 A0A182K4T4 A0A182Q7A6 A0A182VXV1 A0A2P8Y8U6 A0A2M4CYI4 J3JZ30 A0A151I8K0 A0A1L8DW11 A0A0P4VL02 A0A0A9WY87 E2BW95 A0A0A9YXC4 U5EZ35 E2ALZ0 A0A0T6AYV2 T1HUY8 A0A0V0G7A0 A0A0K8U5V0 A0A1I8PZ29 A0A1B0GGE7 A0A1A9ZMI1 A0A224YU72 A0A131YCB8 A0A034WGR1 A0A210QMS3 A0A1I8MRY9 W8BU74 A0A023FFN8 A0A1A9WR99 A0A1S3J6A9 A0A087TI67 K7IWE6 A0A232FD88 B4MM12 B4KXE5 A0A0K8RMG3 A0A0L8GRT4 B4IX16 R7TL43 A0A336LPP1 V4B413
Pubmed
19121390
26354079
22118469
22651552
23622113
18362917
+ More
19820115 26369729 24508170 30249741 28004739 17510324 24945155 24438588 21282665 25244985 12364791 20966253 21347285 21719571 20920257 29403074 22516182 27129103 25401762 20798317 26823975 28797301 26830274 25348373 28812685 25315136 24495485 20075255 28648823 17994087 18057021 23254933
19820115 26369729 24508170 30249741 28004739 17510324 24945155 24438588 21282665 25244985 12364791 20966253 21347285 21719571 20920257 29403074 22516182 27129103 25401762 20798317 26823975 28797301 26830274 25348373 28812685 25315136 24495485 20075255 28648823 17994087 18057021 23254933
EMBL
BABH01002524
DQ443252
ABF51341.1
NWSH01002972
PCG67171.1
GDQN01002815
+ More
JAT88239.1 KQ459602 KPI93358.1 AGBW02009983 OWR49415.1 AK403775 BAM19927.1 KQ460615 KPJ13605.1 GAIX01003315 JAA89245.1 RSAL01000237 RVE43706.1 KQ971343 EFA03370.1 GDAI01000399 JAI17204.1 KK107063 QOIP01000005 EZA60996.1 RLU22766.1 KQ434806 KZC06121.1 GEZM01054608 JAV73527.1 KZ288282 PBC29541.1 KQ435727 KOX77841.1 GFDF01003927 JAV10157.1 GEDC01009655 JAS27643.1 CH477331 EAT43333.1 GFDL01009868 JAV25177.1 GAPW01004324 GEHC01000370 JAC09274.1 JAV47275.1 NEVH01015354 PNF26793.1 ATLV01013778 KE524891 KFB38168.1 DS231931 EDS27320.1 AXCM01002150 GL768061 EFZ12665.1 AJVK01028174 APCN01003116 AAAB01008844 GECU01022222 JAS85484.1 GGFK01011562 MBW44883.1 KQ976574 KYM80141.1 ADTU01015336 KQ981387 KYN42013.1 GECZ01029681 JAS40088.1 LKEY01014849 KYN50499.1 KQ982294 KYQ58300.1 GGFJ01009350 MBW58491.1 GECU01033348 GECU01023893 JAS74358.1 JAS83813.1 KQ414584 KOC70583.1 GL888525 EGI59847.1 GEBQ01016432 JAT23545.1 GGFL01003321 MBW67499.1 AXCN02001147 PYGN01000800 PSN40671.1 GGFL01006218 MBW70396.1 BT128511 AEE63468.1 KQ978346 KYM94756.1 GFDF01003462 JAV10622.1 GDKW01000804 JAI55791.1 GBHO01031218 GBHO01027959 GBHO01006738 GBRD01005547 JAG12386.1 JAG15645.1 JAG36866.1 JAG60274.1 GL451103 EFN79999.1 GBHO01006735 GDHC01021172 JAG36869.1 JAP97456.1 GANO01001504 JAB58367.1 GL440703 EFN65510.1 LJIG01022524 KRT80123.1 ACPB03002116 GECL01002137 JAP03987.1 GDHF01030272 JAI22042.1 CCAG010020395 GFPF01008143 MAA19289.1 GEDV01011583 JAP76974.1 GAKP01005438 GAKP01005435 JAC53517.1 NEDP02002776 OWF50034.1 GAMC01001641 GAMC01001640 JAC04916.1 GBBK01003691 JAC20791.1 KK115333 KFM64806.1 NNAY01000410 OXU28602.1 CH963847 EDW73021.1 CH933809 EDW18631.1 KRG06293.1 GADI01002109 JAA71699.1 KQ420662 KOF79678.1 CH916366 EDV96322.1 AMQN01002527 KB309449 ELT94374.1 UFQT01000097 SSX19755.1 KB203771 ESO83159.1
JAT88239.1 KQ459602 KPI93358.1 AGBW02009983 OWR49415.1 AK403775 BAM19927.1 KQ460615 KPJ13605.1 GAIX01003315 JAA89245.1 RSAL01000237 RVE43706.1 KQ971343 EFA03370.1 GDAI01000399 JAI17204.1 KK107063 QOIP01000005 EZA60996.1 RLU22766.1 KQ434806 KZC06121.1 GEZM01054608 JAV73527.1 KZ288282 PBC29541.1 KQ435727 KOX77841.1 GFDF01003927 JAV10157.1 GEDC01009655 JAS27643.1 CH477331 EAT43333.1 GFDL01009868 JAV25177.1 GAPW01004324 GEHC01000370 JAC09274.1 JAV47275.1 NEVH01015354 PNF26793.1 ATLV01013778 KE524891 KFB38168.1 DS231931 EDS27320.1 AXCM01002150 GL768061 EFZ12665.1 AJVK01028174 APCN01003116 AAAB01008844 GECU01022222 JAS85484.1 GGFK01011562 MBW44883.1 KQ976574 KYM80141.1 ADTU01015336 KQ981387 KYN42013.1 GECZ01029681 JAS40088.1 LKEY01014849 KYN50499.1 KQ982294 KYQ58300.1 GGFJ01009350 MBW58491.1 GECU01033348 GECU01023893 JAS74358.1 JAS83813.1 KQ414584 KOC70583.1 GL888525 EGI59847.1 GEBQ01016432 JAT23545.1 GGFL01003321 MBW67499.1 AXCN02001147 PYGN01000800 PSN40671.1 GGFL01006218 MBW70396.1 BT128511 AEE63468.1 KQ978346 KYM94756.1 GFDF01003462 JAV10622.1 GDKW01000804 JAI55791.1 GBHO01031218 GBHO01027959 GBHO01006738 GBRD01005547 JAG12386.1 JAG15645.1 JAG36866.1 JAG60274.1 GL451103 EFN79999.1 GBHO01006735 GDHC01021172 JAG36869.1 JAP97456.1 GANO01001504 JAB58367.1 GL440703 EFN65510.1 LJIG01022524 KRT80123.1 ACPB03002116 GECL01002137 JAP03987.1 GDHF01030272 JAI22042.1 CCAG010020395 GFPF01008143 MAA19289.1 GEDV01011583 JAP76974.1 GAKP01005438 GAKP01005435 JAC53517.1 NEDP02002776 OWF50034.1 GAMC01001641 GAMC01001640 JAC04916.1 GBBK01003691 JAC20791.1 KK115333 KFM64806.1 NNAY01000410 OXU28602.1 CH963847 EDW73021.1 CH933809 EDW18631.1 KRG06293.1 GADI01002109 JAA71699.1 KQ420662 KOF79678.1 CH916366 EDV96322.1 AMQN01002527 KB309449 ELT94374.1 UFQT01000097 SSX19755.1 KB203771 ESO83159.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000283053
+ More
UP000007266 UP000053097 UP000279307 UP000192223 UP000005203 UP000076502 UP000242457 UP000053105 UP000008820 UP000235965 UP000075880 UP000030765 UP000002320 UP000075883 UP000076408 UP000092462 UP000075840 UP000075885 UP000075903 UP000075900 UP000076407 UP000075882 UP000075902 UP000075884 UP000069272 UP000078540 UP000005205 UP000078541 UP000078492 UP000075809 UP000053825 UP000007755 UP000075881 UP000075886 UP000075920 UP000245037 UP000078542 UP000008237 UP000000311 UP000015103 UP000095300 UP000092444 UP000092445 UP000242188 UP000095301 UP000091820 UP000085678 UP000054359 UP000002358 UP000215335 UP000007798 UP000009192 UP000053454 UP000001070 UP000014760 UP000030746
UP000007266 UP000053097 UP000279307 UP000192223 UP000005203 UP000076502 UP000242457 UP000053105 UP000008820 UP000235965 UP000075880 UP000030765 UP000002320 UP000075883 UP000076408 UP000092462 UP000075840 UP000075885 UP000075903 UP000075900 UP000076407 UP000075882 UP000075902 UP000075884 UP000069272 UP000078540 UP000005205 UP000078541 UP000078492 UP000075809 UP000053825 UP000007755 UP000075881 UP000075886 UP000075920 UP000245037 UP000078542 UP000008237 UP000000311 UP000015103 UP000095300 UP000092444 UP000092445 UP000242188 UP000095301 UP000091820 UP000085678 UP000054359 UP000002358 UP000215335 UP000007798 UP000009192 UP000053454 UP000001070 UP000014760 UP000030746
Interpro
SUPFAM
SSF47694
SSF47694
Gene 3D
ProteinModelPortal
H9JE31
Q1HQ00
A0A2A4J5B7
A0A1E1WMJ1
A0A194PK09
A0A212F6Q2
+ More
I4DPT7 A0A194R8N1 S4PMX3 A0A3S2LTP6 D6WM18 A0A0K8TS74 A0A026WZ20 A0A1W4X3S9 A0A088AM34 A0A154P2L7 A0A1Y1LR73 A0A2A3EEV0 A0A0N0BIR7 A0A1L8DUP4 A0A1B6DPV0 Q17AQ3 A0A1S4F9W4 A0A1Q3FC92 A0A023ELX1 A0A2J7QDZ7 A0A182J0I4 A0A084VJM4 B0WGZ5 A0A182M6V1 E9J3D6 A0A182XXE0 A0A1B0D9D3 A0A182I8T3 A0A182P786 A0A182UVX7 A0A182R5X2 A0A182WV58 A0A1S4GJ96 A0A182LAP9 A0A182UA30 A0A1Y9H2L8 A0A1B6IEY5 A0A2M4AVR4 A0A1I8JSN3 A0A195B6K8 A0A158NGY9 A0A195FNG0 A0A1B6EQ79 A0A151K2Z8 A0A151XD97 A0A2M4C032 A0A1B6IA95 A0A0L7RIF6 F4X136 A0A1B6LIN1 A0A087YXN2 A0A2M4CQE7 A0A182K4T4 A0A182Q7A6 A0A182VXV1 A0A2P8Y8U6 A0A2M4CYI4 J3JZ30 A0A151I8K0 A0A1L8DW11 A0A0P4VL02 A0A0A9WY87 E2BW95 A0A0A9YXC4 U5EZ35 E2ALZ0 A0A0T6AYV2 T1HUY8 A0A0V0G7A0 A0A0K8U5V0 A0A1I8PZ29 A0A1B0GGE7 A0A1A9ZMI1 A0A224YU72 A0A131YCB8 A0A034WGR1 A0A210QMS3 A0A1I8MRY9 W8BU74 A0A023FFN8 A0A1A9WR99 A0A1S3J6A9 A0A087TI67 K7IWE6 A0A232FD88 B4MM12 B4KXE5 A0A0K8RMG3 A0A0L8GRT4 B4IX16 R7TL43 A0A336LPP1 V4B413
I4DPT7 A0A194R8N1 S4PMX3 A0A3S2LTP6 D6WM18 A0A0K8TS74 A0A026WZ20 A0A1W4X3S9 A0A088AM34 A0A154P2L7 A0A1Y1LR73 A0A2A3EEV0 A0A0N0BIR7 A0A1L8DUP4 A0A1B6DPV0 Q17AQ3 A0A1S4F9W4 A0A1Q3FC92 A0A023ELX1 A0A2J7QDZ7 A0A182J0I4 A0A084VJM4 B0WGZ5 A0A182M6V1 E9J3D6 A0A182XXE0 A0A1B0D9D3 A0A182I8T3 A0A182P786 A0A182UVX7 A0A182R5X2 A0A182WV58 A0A1S4GJ96 A0A182LAP9 A0A182UA30 A0A1Y9H2L8 A0A1B6IEY5 A0A2M4AVR4 A0A1I8JSN3 A0A195B6K8 A0A158NGY9 A0A195FNG0 A0A1B6EQ79 A0A151K2Z8 A0A151XD97 A0A2M4C032 A0A1B6IA95 A0A0L7RIF6 F4X136 A0A1B6LIN1 A0A087YXN2 A0A2M4CQE7 A0A182K4T4 A0A182Q7A6 A0A182VXV1 A0A2P8Y8U6 A0A2M4CYI4 J3JZ30 A0A151I8K0 A0A1L8DW11 A0A0P4VL02 A0A0A9WY87 E2BW95 A0A0A9YXC4 U5EZ35 E2ALZ0 A0A0T6AYV2 T1HUY8 A0A0V0G7A0 A0A0K8U5V0 A0A1I8PZ29 A0A1B0GGE7 A0A1A9ZMI1 A0A224YU72 A0A131YCB8 A0A034WGR1 A0A210QMS3 A0A1I8MRY9 W8BU74 A0A023FFN8 A0A1A9WR99 A0A1S3J6A9 A0A087TI67 K7IWE6 A0A232FD88 B4MM12 B4KXE5 A0A0K8RMG3 A0A0L8GRT4 B4IX16 R7TL43 A0A336LPP1 V4B413
Ontologies
KEGG
GO
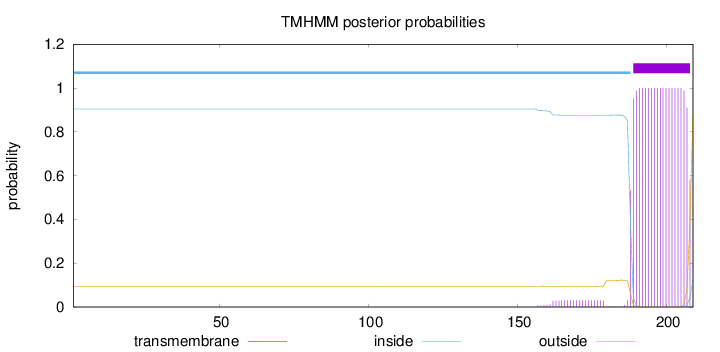
Topology
Length:
209
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.58294
Exp number, first 60 AAs:
0
Total prob of N-in:
0.90475
inside
1 - 188
TMhelix
189 - 208
outside
209 - 209
Population Genetic Test Statistics
Pi
276.785427
Theta
178.109842
Tajima's D
1.669063
CLR
0.003529
CSRT
0.826108694565272
Interpretation
Uncertain
