Gene
KWMTBOMO08998
Annotation
PREDICTED:_pseudouridine-metabolizing_bifunctional_protein_C1861.05_isoform_X3_[Bombyx_mori]
Full name
Pseudouridine-5'-phosphate glycosidase
Location in the cell
Mitochondrial Reliability : 2.046
Sequence
CDS
ATGGCGTACCGACTAAATGGCATACCGAAACTTGTTCAGCGTAATTTGGGTTTTCTTAGAAATTATAGCGAGTCTCGTCATCCCTTCGTTTATTCCAAGGAAGTTTCTCGGGCGAAGTCAGAAAATATGCCAATAGTAGCCCTCGAATCAACTATTATTACACACGGAATGCCTTACCCACAGAACTTGGAAACGGCTCTGGAAGTGGAGAATATAATTAGGCAACGGGGTGCCGTTCCAGCGACTGTGGCAATTCTTAAAGGGCAATTGACCGTTGGTCTGACGGAAGATCAACTTCGATACCTTGCTCAGGCCAAAGGTGTAATTAAAGCATCGAGACGTGATCTTGCTTATATAGCTGCGGCTAAGCTTGACGGTGCCACAACCGTCGCTGGGACAATTATTGCTTCAGAATTAGCTGACATACCAGTCTTCGTGACCGGTGGAATTGGGGGCGTCCATCGTGAAGGTGAAAGTACACTAGACATCTCAGCAGATCTGAATGAACTCGGGCGAAGCAAAACACTCGTTGTTTGCAGCGGAATCAAATCAATACTCGACATTGGAAGAACTCTCGAATATTTGGAAACGCAAGGCGTCTGTGTGTGTTCGTTTGGCGAGTCTAACGAGTTTCCAGCATTTTACACGGTCCGTTCCGGTCACCGGGCTCCGTATAGCGTTGCCGACGCGAAGCACGCCGCTCGGATTCTTCACGAATCACACAGATTCCACCTCAATTCCGGCGTCGTCATCGCTGTTCCTGTGCCGCGACGTGACGCTATGGACGAAAAAGTAATTGAAGAGGCTATCAATAGTGCCCTGACGGATGCCAAAAGAAAAGGTATTCGCGGTAAAGAAGTTACTCCTTACATTTTATCGAACGTCAGTGAAGCGACCAGCGGCACGTCACTTGAAACCAATATCGCATTGATAAAGAACAATGCGCGCGTCGGCGCCGATATAGCCGTCGAATTTAAAAAACTCAAGAACGCCGACAATGTAAACGATTCTAATATTGGATTCCGCAAGGGCGCCGAGAATATCTCGAATTTCACGAGGACATTTCACACTTCGTCTAATACGAGGAGCGGGGCGGAGAATTGCGGAGATGATACGCCTATGCTCGCTTTAGATATGAAGGCGGACGGAGACGTGCTCGTGATCGGAGGGGCCAACGTTGACAGAACCTACAGAGTATCAGAAGATACCATTCAGCACAATTATGTTCGCGTGTCCGAGCGAGACAGCGAGATATTGCTTCTTCACACCAACTCGCACTGTTGA
Protein
MAYRLNGIPKLVQRNLGFLRNYSESRHPFVYSKEVSRAKSENMPIVALESTIITHGMPYPQNLETALEVENIIRQRGAVPATVAILKGQLTVGLTEDQLRYLAQAKGVIKASRRDLAYIAAAKLDGATTVAGTIIASELADIPVFVTGGIGGVHREGESTLDISADLNELGRSKTLVVCSGIKSILDIGRTLEYLETQGVCVCSFGESNEFPAFYTVRSGHRAPYSVADAKHAARILHESHRFHLNSGVVIAVPVPRRDAMDEKVIEEAINSALTDAKRKGIRGKEVTPYILSNVSEATSGTSLETNIALIKNNARVGADIAVEFKKLKNADNVNDSNIGFRKGAENISNFTRTFHTSSNTRSGAENCGDDTPMLALDMKADGDVLVIGGANVDRTYRVSEDTIQHNYVRVSERDSEILLLHTNSHC
Summary
Description
Catalyzes the reversible cleavage of pseudouridine 5'-phosphate (PsiMP) to ribose 5-phosphate and uracil. Functions biologically in the cleavage direction, as part of a pseudouridine degradation pathway.
Catalytic Activity
D-ribose 5-phosphate + uracil = H2O + psi-UMP
Cofactor
Mn(2+)
Subunit
Homotrimer.
Similarity
Belongs to the pseudouridine-5'-phosphate glycosidase family.
Belongs to the carbohydrate kinase PfkB family.
Belongs to the carbohydrate kinase PfkB family.
Uniprot
H9JE33
A0A2H1WUY8
A0A2A4JUW0
A0A194PK04
A0A194R704
A0A212F6P5
+ More
A0A452HRQ7 A0A1B6J3W4 A0A1B6K1A6 A0A084VBP9 H0ZN49 A0A1I6RYZ6 A0A182INK8 A0A3L8SGV1 A0A218V0X4 A0A182VIA2 A0A3Q3T1L6 A0A0P6GLY1 A0A1L8GW64 A0A1W4W8Z3 A0A285CS70 A0A0C7B735 A0A367J1X1 A0A0P5KGR8 A0A1J1IF73 A0A1X0RM47 A0A164XRG1 A0A182F9G3 A0A231RY48 A0A0P5DBB6 A0A1U6HKE9 A0A0P5QIH9 A0A0P5KM36 A0A0P6DCF7 A0A0N8BDK1 A0A0P5SHL0 A0A0P5LFI6 A0A0P5M4C4 A0A1J6WRK1 A0A0P4ZRU7 A0A0P5NMP2 A0A0P6WVQ5 A0A0P5GDW0 A0A0P5NZJ3 A0A0P5DKZ8 A0A0N8CFC0 A0A0P5J066 A0A0P5D6K7 A0A1B6BE42 A0A0P5LIS2 A0A0C1R7J9 A0A0P5LDJ7 A0A0P5W0Y9 A0A0P5JWV2 A0A0P6DWD4 A0A1A8CU46 A0A0P5ITJ9 A0A0P5WWE3 A0A0P5D2R0 A0A182XXD4 A0A0P5A5C8 A0A367JCP8 E9HD67 A0A1M5S1Q6 A0A068RTJ3 A0A3B4XWJ6 A0A0P4ZBZ9 A0A2D0RHN7 A0A2G4T858 A0A1X0RFX8 A0A0N8ARS7
A0A452HRQ7 A0A1B6J3W4 A0A1B6K1A6 A0A084VBP9 H0ZN49 A0A1I6RYZ6 A0A182INK8 A0A3L8SGV1 A0A218V0X4 A0A182VIA2 A0A3Q3T1L6 A0A0P6GLY1 A0A1L8GW64 A0A1W4W8Z3 A0A285CS70 A0A0C7B735 A0A367J1X1 A0A0P5KGR8 A0A1J1IF73 A0A1X0RM47 A0A164XRG1 A0A182F9G3 A0A231RY48 A0A0P5DBB6 A0A1U6HKE9 A0A0P5QIH9 A0A0P5KM36 A0A0P6DCF7 A0A0N8BDK1 A0A0P5SHL0 A0A0P5LFI6 A0A0P5M4C4 A0A1J6WRK1 A0A0P4ZRU7 A0A0P5NMP2 A0A0P6WVQ5 A0A0P5GDW0 A0A0P5NZJ3 A0A0P5DKZ8 A0A0N8CFC0 A0A0P5J066 A0A0P5D6K7 A0A1B6BE42 A0A0P5LIS2 A0A0C1R7J9 A0A0P5LDJ7 A0A0P5W0Y9 A0A0P5JWV2 A0A0P6DWD4 A0A1A8CU46 A0A0P5ITJ9 A0A0P5WWE3 A0A0P5D2R0 A0A182XXD4 A0A0P5A5C8 A0A367JCP8 E9HD67 A0A1M5S1Q6 A0A068RTJ3 A0A3B4XWJ6 A0A0P4ZBZ9 A0A2D0RHN7 A0A2G4T858 A0A1X0RFX8 A0A0N8ARS7
EC Number
4.2.1.70
Pubmed
EMBL
BABH01002526
ODYU01011263
SOQ56889.1
NWSH01000600
PCG75414.1
KQ459602
+ More
KPI93353.1 KQ460615 KPJ13598.1 AGBW02009983 OWR49412.1 GECU01037450 GECU01013842 JAS70256.1 JAS93864.1 GECU01002470 JAT05237.1 ATLV01008987 KE524494 KFB35393.1 ABQF01016418 ABQF01016419 FPAA01000006 SFS69896.1 QUSF01000021 RLW01861.1 MUZQ01000075 OWK59576.1 GDIQ01043501 JAN51236.1 CM004470 OCT88087.1 OAOP01000004 SNX70407.1 CCYT01000006 CEG66759.1 PJQL01002543 RCH83829.1 GDIQ01191140 JAK60585.1 CVRI01000048 CRK98919.1 KV921571 ORE13077.1 LRGB01000944 KZS14499.1 MWSH01000001 OXS64367.1 GDIP01174734 JAJ48668.1 PVYY01000001 FVZH01000001 PRX79505.1 SLJ96284.1 GDIQ01113682 JAL38044.1 GDIQ01189598 JAK62127.1 GDIQ01079247 JAN15490.1 GDIQ01188251 JAK63474.1 GDIP01139644 JAL64070.1 GDIQ01170362 JAK81363.1 GDIQ01169566 JAK82159.1 MINN01000106 OIU70535.1 GDIP01208812 JAJ14590.1 GDIQ01144233 JAL07493.1 LIXZ01000003 KPL60501.1 GDIQ01243940 JAK07785.1 GDIQ01145626 JAL06100.1 GDIP01155791 JAJ67611.1 GDIP01137287 JAL66427.1 GDIQ01208209 JAK43516.1 GDIP01160484 JAJ62918.1 BDHH01000010 GAU77991.1 GDIQ01173007 JAK78718.1 AYSO01000017 KIE46491.1 GDIQ01173006 JAK78719.1 GDIP01093286 JAM10429.1 GDIQ01197854 JAK53871.1 GDIQ01071285 JAN23452.1 HADZ01018280 SBP82221.1 GDIQ01208210 JAK43515.1 GDIP01080638 JAM23077.1 GDIP01162075 JAJ61327.1 GDIP01203829 JAJ19573.1 PJQL01001603 RCH87718.1 GL732622 EFX70313.1 FQXH01000016 SHH32379.1 CBTN010000017 CDH53349.1 GDIP01215380 JAJ08022.1 KZ303842 PHZ16856.1 KV921861 ORE10945.1 GDIQ01249243 JAK02482.1
KPI93353.1 KQ460615 KPJ13598.1 AGBW02009983 OWR49412.1 GECU01037450 GECU01013842 JAS70256.1 JAS93864.1 GECU01002470 JAT05237.1 ATLV01008987 KE524494 KFB35393.1 ABQF01016418 ABQF01016419 FPAA01000006 SFS69896.1 QUSF01000021 RLW01861.1 MUZQ01000075 OWK59576.1 GDIQ01043501 JAN51236.1 CM004470 OCT88087.1 OAOP01000004 SNX70407.1 CCYT01000006 CEG66759.1 PJQL01002543 RCH83829.1 GDIQ01191140 JAK60585.1 CVRI01000048 CRK98919.1 KV921571 ORE13077.1 LRGB01000944 KZS14499.1 MWSH01000001 OXS64367.1 GDIP01174734 JAJ48668.1 PVYY01000001 FVZH01000001 PRX79505.1 SLJ96284.1 GDIQ01113682 JAL38044.1 GDIQ01189598 JAK62127.1 GDIQ01079247 JAN15490.1 GDIQ01188251 JAK63474.1 GDIP01139644 JAL64070.1 GDIQ01170362 JAK81363.1 GDIQ01169566 JAK82159.1 MINN01000106 OIU70535.1 GDIP01208812 JAJ14590.1 GDIQ01144233 JAL07493.1 LIXZ01000003 KPL60501.1 GDIQ01243940 JAK07785.1 GDIQ01145626 JAL06100.1 GDIP01155791 JAJ67611.1 GDIP01137287 JAL66427.1 GDIQ01208209 JAK43516.1 GDIP01160484 JAJ62918.1 BDHH01000010 GAU77991.1 GDIQ01173007 JAK78718.1 AYSO01000017 KIE46491.1 GDIQ01173006 JAK78719.1 GDIP01093286 JAM10429.1 GDIQ01197854 JAK53871.1 GDIQ01071285 JAN23452.1 HADZ01018280 SBP82221.1 GDIQ01208210 JAK43515.1 GDIP01080638 JAM23077.1 GDIP01162075 JAJ61327.1 GDIP01203829 JAJ19573.1 PJQL01001603 RCH87718.1 GL732622 EFX70313.1 FQXH01000016 SHH32379.1 CBTN010000017 CDH53349.1 GDIP01215380 JAJ08022.1 KZ303842 PHZ16856.1 KV921861 ORE10945.1 GDIQ01249243 JAK02482.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000291020
+ More
UP000030765 UP000007754 UP000198660 UP000075880 UP000276834 UP000197619 UP000075903 UP000261640 UP000186698 UP000192223 UP000219546 UP000054919 UP000252139 UP000183832 UP000242381 UP000076858 UP000069272 UP000214946 UP000189895 UP000237918 UP000182062 UP000050398 UP000095197 UP000031366 UP000076408 UP000000305 UP000242520 UP000027586 UP000261360 UP000221080 UP000242254 UP000242414
UP000030765 UP000007754 UP000198660 UP000075880 UP000276834 UP000197619 UP000075903 UP000261640 UP000186698 UP000192223 UP000219546 UP000054919 UP000252139 UP000183832 UP000242381 UP000076858 UP000069272 UP000214946 UP000189895 UP000237918 UP000182062 UP000050398 UP000095197 UP000031366 UP000076408 UP000000305 UP000242520 UP000027586 UP000261360 UP000221080 UP000242254 UP000242414
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JE33
A0A2H1WUY8
A0A2A4JUW0
A0A194PK04
A0A194R704
A0A212F6P5
+ More
A0A452HRQ7 A0A1B6J3W4 A0A1B6K1A6 A0A084VBP9 H0ZN49 A0A1I6RYZ6 A0A182INK8 A0A3L8SGV1 A0A218V0X4 A0A182VIA2 A0A3Q3T1L6 A0A0P6GLY1 A0A1L8GW64 A0A1W4W8Z3 A0A285CS70 A0A0C7B735 A0A367J1X1 A0A0P5KGR8 A0A1J1IF73 A0A1X0RM47 A0A164XRG1 A0A182F9G3 A0A231RY48 A0A0P5DBB6 A0A1U6HKE9 A0A0P5QIH9 A0A0P5KM36 A0A0P6DCF7 A0A0N8BDK1 A0A0P5SHL0 A0A0P5LFI6 A0A0P5M4C4 A0A1J6WRK1 A0A0P4ZRU7 A0A0P5NMP2 A0A0P6WVQ5 A0A0P5GDW0 A0A0P5NZJ3 A0A0P5DKZ8 A0A0N8CFC0 A0A0P5J066 A0A0P5D6K7 A0A1B6BE42 A0A0P5LIS2 A0A0C1R7J9 A0A0P5LDJ7 A0A0P5W0Y9 A0A0P5JWV2 A0A0P6DWD4 A0A1A8CU46 A0A0P5ITJ9 A0A0P5WWE3 A0A0P5D2R0 A0A182XXD4 A0A0P5A5C8 A0A367JCP8 E9HD67 A0A1M5S1Q6 A0A068RTJ3 A0A3B4XWJ6 A0A0P4ZBZ9 A0A2D0RHN7 A0A2G4T858 A0A1X0RFX8 A0A0N8ARS7
A0A452HRQ7 A0A1B6J3W4 A0A1B6K1A6 A0A084VBP9 H0ZN49 A0A1I6RYZ6 A0A182INK8 A0A3L8SGV1 A0A218V0X4 A0A182VIA2 A0A3Q3T1L6 A0A0P6GLY1 A0A1L8GW64 A0A1W4W8Z3 A0A285CS70 A0A0C7B735 A0A367J1X1 A0A0P5KGR8 A0A1J1IF73 A0A1X0RM47 A0A164XRG1 A0A182F9G3 A0A231RY48 A0A0P5DBB6 A0A1U6HKE9 A0A0P5QIH9 A0A0P5KM36 A0A0P6DCF7 A0A0N8BDK1 A0A0P5SHL0 A0A0P5LFI6 A0A0P5M4C4 A0A1J6WRK1 A0A0P4ZRU7 A0A0P5NMP2 A0A0P6WVQ5 A0A0P5GDW0 A0A0P5NZJ3 A0A0P5DKZ8 A0A0N8CFC0 A0A0P5J066 A0A0P5D6K7 A0A1B6BE42 A0A0P5LIS2 A0A0C1R7J9 A0A0P5LDJ7 A0A0P5W0Y9 A0A0P5JWV2 A0A0P6DWD4 A0A1A8CU46 A0A0P5ITJ9 A0A0P5WWE3 A0A0P5D2R0 A0A182XXD4 A0A0P5A5C8 A0A367JCP8 E9HD67 A0A1M5S1Q6 A0A068RTJ3 A0A3B4XWJ6 A0A0P4ZBZ9 A0A2D0RHN7 A0A2G4T858 A0A1X0RFX8 A0A0N8ARS7
PDB
4GIL
E-value=2.40761e-70,
Score=675
Ontologies
PATHWAY
GO
PANTHER
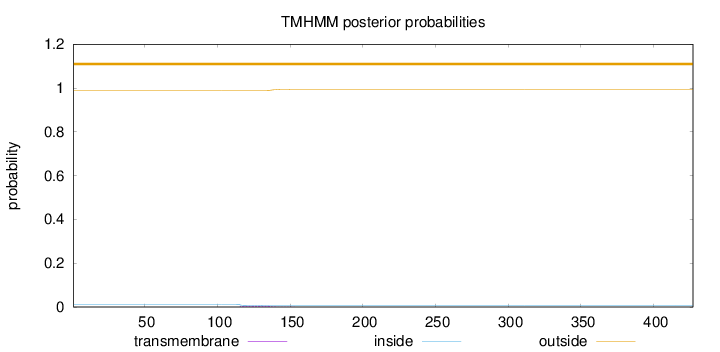
Topology
Length:
427
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12511
Exp number, first 60 AAs:
0.00072
Total prob of N-in:
0.01216
outside
1 - 427
Population Genetic Test Statistics
Pi
249.111036
Theta
161.278586
Tajima's D
1.67282
CLR
0.273276
CSRT
0.822858857057147
Interpretation
Uncertain
