Pre Gene Modal
BGIBMGA007781
Annotation
PREDICTED:_pseudouridine-metabolizing_bifunctional_protein_C1861.05_isoform_X3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.35 Mitochondrial Reliability : 1.185
Sequence
CDS
ATGTTGGACGGCAGTACACATGCTTGCGTCATGGATCAATGCGGCGGTGGAGTCGGACGTAACATGGCAGAAGCACTATGGAGACTGCGGGGCGGACGTACGAAACTTCTGACCGCCATAGGCGACGATGCCGATGGAAAATACTTAGAAGAGATTGCACCAGGCCTGATACTGAACGGATGCGTCGTAAAGAACGGTCGGACGCCAACATATGCGGCAGTGATGGATTCCCGGGGAGAATGCCTTCTAGGTCTCGGCGACATGGCATTACACAATGACATCACCGTCGACAGGGTAAACAATAATATGACGCTATTAGAAAGGGCTCCACTTGTAGTTCTCGATGGCAACGTCCCACAAATTACTATGGACCATGTCTTAGAGCAATGTCAACGGCTTCGAAAACCTGGTAAGTTTCTAGAACGCTCTTGA
Protein
MLDGSTHACVMDQCGGGVGRNMAEALWRLRGGRTKLLTAIGDDADGKYLEEIAPGLILNGCVVKNGRTPTYAAVMDSRGECLLGLGDMALHNDITVDRVNNNMTLLERAPLVVLDGNVPQITMDHVLEQCQRLRKPGKFLERS
Summary
Uniprot
H9JE34
A0A2H1WGX3
A0A2A4J5P7
A0A194R8M2
A0A194PQE0
A0A3S2NUB5
+ More
A0A212F6P9 A0A2P8YK20 A0A293LJT0 A0A293LIA6 B0WVP4 A0A1D2MGV0 Q17AQ9 A0A182NFW6 A0A182XXD0 B4KFG8 A0A0J9QU64 B4Q657 B4ICY9 Q9VPR3 Q8MYS6 B4LTN4 B3N7S1 A0A0M4E876 A0A0P5GTV1 A0A0P5LIS2 A0A0P5A5C8 A0A0P5LDJ7 A0A0P4ZBZ9 B4P2G1 A0A0P5JMQ7 A0A147BGQ2 A0A182I8S2 A0A1W4V2M9 A0A164XRJ0 B4JDR4 A0A167U815 A0A0P5WWE3 J9M3P9 A0A0P5W0Y9 A0A0N8ARS7 A0A0P5D2R0 A0A0P6GFA6 A0A0P6HQP4 A0A0G3XYQ0 A0A063YYC2 A0A023CLX3 L7ZQQ7 A0A1J1IFD5 A0A0P5A7V9 A0A0P6D064 S5YYB4 A0A0E0TAL8 A0A087LEC1 A0A161SHX7 A0A3L7CZ22 A0A2M9T249 A0A0D8BQU5 A0A1V9B3H9 A0A1B6D3E9 A0A1B6DRE2 A0A210PPV3 B3MLP7 A0A377FZG5 A0A182MJW6 A0A150M7Y9 A0A3B0JHG4 A0A142CZS0 V6VCM7 A0A2M4DS25 A0A0P5ZSC8 A0A2M4CRL9 Q29N57 B4G882 A0A1V9C1A7 A0A0Q0Y9E8 A0A150NF66 A0A3S0IXZ4 A0A3B0UKN6 A0A1I8PQD8 A0A1A9WN01 A0A139WH26 A0A3Q9QS05 A0A250DYF0 A0A2S2R297 A0A1L8E1E1 A0A1V4P819 A0A1Q5SR52 U2X8P4 H2J7F3 A0A069DYJ8 R9MSM4 A0A0N0Z1I2 A0A1A9XXR0 A0A1B0APG1 A0A1J4THC7 A0A1A9Z630 A0A226Q1I4 A0A0J0V357 A0A093IYC1
A0A212F6P9 A0A2P8YK20 A0A293LJT0 A0A293LIA6 B0WVP4 A0A1D2MGV0 Q17AQ9 A0A182NFW6 A0A182XXD0 B4KFG8 A0A0J9QU64 B4Q657 B4ICY9 Q9VPR3 Q8MYS6 B4LTN4 B3N7S1 A0A0M4E876 A0A0P5GTV1 A0A0P5LIS2 A0A0P5A5C8 A0A0P5LDJ7 A0A0P4ZBZ9 B4P2G1 A0A0P5JMQ7 A0A147BGQ2 A0A182I8S2 A0A1W4V2M9 A0A164XRJ0 B4JDR4 A0A167U815 A0A0P5WWE3 J9M3P9 A0A0P5W0Y9 A0A0N8ARS7 A0A0P5D2R0 A0A0P6GFA6 A0A0P6HQP4 A0A0G3XYQ0 A0A063YYC2 A0A023CLX3 L7ZQQ7 A0A1J1IFD5 A0A0P5A7V9 A0A0P6D064 S5YYB4 A0A0E0TAL8 A0A087LEC1 A0A161SHX7 A0A3L7CZ22 A0A2M9T249 A0A0D8BQU5 A0A1V9B3H9 A0A1B6D3E9 A0A1B6DRE2 A0A210PPV3 B3MLP7 A0A377FZG5 A0A182MJW6 A0A150M7Y9 A0A3B0JHG4 A0A142CZS0 V6VCM7 A0A2M4DS25 A0A0P5ZSC8 A0A2M4CRL9 Q29N57 B4G882 A0A1V9C1A7 A0A0Q0Y9E8 A0A150NF66 A0A3S0IXZ4 A0A3B0UKN6 A0A1I8PQD8 A0A1A9WN01 A0A139WH26 A0A3Q9QS05 A0A250DYF0 A0A2S2R297 A0A1L8E1E1 A0A1V4P819 A0A1Q5SR52 U2X8P4 H2J7F3 A0A069DYJ8 R9MSM4 A0A0N0Z1I2 A0A1A9XXR0 A0A1B0APG1 A0A1J4THC7 A0A1A9Z630 A0A226Q1I4 A0A0J0V357 A0A093IYC1
Pubmed
EMBL
BABH01002532
ODYU01008491
SOQ52142.1
NWSH01003076
PCG66998.1
KQ460615
+ More
KPJ13595.1 KQ459602 KPI93350.1 RSAL01000187 RVE44770.1 AGBW02009983 OWR49410.1 PYGN01000538 PSN44599.1 GFWV01002340 MAA27070.1 GFWV01003733 MAA28463.1 DS232130 EDS35651.1 LJIJ01001329 ODM92094.1 CH477331 EAT43327.1 CH933807 EDW13083.1 CM002910 KMY87339.1 CM000361 EDX03241.1 CH480829 EDW45415.1 AE014134 AAF51480.1 AY113628 AAM29633.1 CH940649 EDW65007.1 CH954177 EDV57247.1 CP012523 ALC38164.1 GDIQ01239840 JAK11885.1 GDIQ01173007 JAK78718.1 GDIP01203829 JAJ19573.1 GDIQ01173006 JAK78719.1 GDIP01215380 JAJ08022.1 CM000157 EDW87157.1 GDIQ01197853 JAK53872.1 GEGO01005466 JAR89938.1 APCN01003108 LRGB01000944 KZS14501.1 CH916368 EDW03434.1 CP015436 ANB64204.1 GDIP01080638 JAM23077.1 ABLF02022385 ABLF02022386 ABLF02022389 ABLF02022390 GDIP01093286 JAM10429.1 GDIQ01249243 JAK02482.1 GDIP01162075 JAJ61327.1 GDIQ01034017 JAN60720.1 GDIQ01016411 JAN78326.1 CP011832 AKM18676.1 JHUS01000030 KDE49317.1 AOTZ01000006 EZP76422.1 CP004008 AGE21833.1 CVRI01000048 CRK98928.1 GDIP01203828 JAJ19574.1 GDIP01007424 JAM96291.1 CP006254 AGT31684.1 CP002442 ADU93753.1 JPYV01000093 KFX34140.1 LQIE01000187 KZE90460.1 RCTJ01000007 RLQ14766.1 PIJF01000036 PJW16970.1 JYBP01000003 KJE26354.1 NADP01000011 OQP03936.1 GEDC01026456 GEDC01017076 JAS10842.1 JAS20222.1 GEDC01009042 JAS28256.1 NEDP02005564 OWF38530.1 CH902620 EDV30768.2 UGGS01000001 STO11891.1 AXCM01001575 LQYV01000152 KYD20465.1 OUUW01000004 SPP79732.1 CP014749 AMQ20272.1 AYSF01000055 ESU71806.1 GGFL01016179 MBW80357.1 GDIP01040299 JAM63416.1 GGFL01003737 MBW67915.1 CH379060 EAL33486.1 CH479180 EDW28562.1 NAGE01000043 OQP15376.1 LLKS01000069 KQC45951.1 LQYY01000002 KYD35367.1 RXNT01000004 RTR33840.1 UOER01000529 VAW25917.1 KQ971343 KYB27263.1 CP022572 AZU60882.1 CP016552 ATA59691.1 GGMS01014903 MBY84106.1 GFDF01001575 JAV12509.1 MVKA01000020 OPX02956.1 MQMG01000046 OKO90396.1 BASG01000057 GAD15192.1 CP003257 AEX86446.1 GBGD01002265 JAC86624.1 ASTD01000106 EOS73676.1 LJAJ01000058 KPC97718.1 JXJN01001399 MNUW01000075 OIO11218.1 CP018058 ASS99797.1 LDNZ01000185 KLR72735.1 KK571601 KFW07745.1
KPJ13595.1 KQ459602 KPI93350.1 RSAL01000187 RVE44770.1 AGBW02009983 OWR49410.1 PYGN01000538 PSN44599.1 GFWV01002340 MAA27070.1 GFWV01003733 MAA28463.1 DS232130 EDS35651.1 LJIJ01001329 ODM92094.1 CH477331 EAT43327.1 CH933807 EDW13083.1 CM002910 KMY87339.1 CM000361 EDX03241.1 CH480829 EDW45415.1 AE014134 AAF51480.1 AY113628 AAM29633.1 CH940649 EDW65007.1 CH954177 EDV57247.1 CP012523 ALC38164.1 GDIQ01239840 JAK11885.1 GDIQ01173007 JAK78718.1 GDIP01203829 JAJ19573.1 GDIQ01173006 JAK78719.1 GDIP01215380 JAJ08022.1 CM000157 EDW87157.1 GDIQ01197853 JAK53872.1 GEGO01005466 JAR89938.1 APCN01003108 LRGB01000944 KZS14501.1 CH916368 EDW03434.1 CP015436 ANB64204.1 GDIP01080638 JAM23077.1 ABLF02022385 ABLF02022386 ABLF02022389 ABLF02022390 GDIP01093286 JAM10429.1 GDIQ01249243 JAK02482.1 GDIP01162075 JAJ61327.1 GDIQ01034017 JAN60720.1 GDIQ01016411 JAN78326.1 CP011832 AKM18676.1 JHUS01000030 KDE49317.1 AOTZ01000006 EZP76422.1 CP004008 AGE21833.1 CVRI01000048 CRK98928.1 GDIP01203828 JAJ19574.1 GDIP01007424 JAM96291.1 CP006254 AGT31684.1 CP002442 ADU93753.1 JPYV01000093 KFX34140.1 LQIE01000187 KZE90460.1 RCTJ01000007 RLQ14766.1 PIJF01000036 PJW16970.1 JYBP01000003 KJE26354.1 NADP01000011 OQP03936.1 GEDC01026456 GEDC01017076 JAS10842.1 JAS20222.1 GEDC01009042 JAS28256.1 NEDP02005564 OWF38530.1 CH902620 EDV30768.2 UGGS01000001 STO11891.1 AXCM01001575 LQYV01000152 KYD20465.1 OUUW01000004 SPP79732.1 CP014749 AMQ20272.1 AYSF01000055 ESU71806.1 GGFL01016179 MBW80357.1 GDIP01040299 JAM63416.1 GGFL01003737 MBW67915.1 CH379060 EAL33486.1 CH479180 EDW28562.1 NAGE01000043 OQP15376.1 LLKS01000069 KQC45951.1 LQYY01000002 KYD35367.1 RXNT01000004 RTR33840.1 UOER01000529 VAW25917.1 KQ971343 KYB27263.1 CP022572 AZU60882.1 CP016552 ATA59691.1 GGMS01014903 MBY84106.1 GFDF01001575 JAV12509.1 MVKA01000020 OPX02956.1 MQMG01000046 OKO90396.1 BASG01000057 GAD15192.1 CP003257 AEX86446.1 GBGD01002265 JAC86624.1 ASTD01000106 EOS73676.1 LJAJ01000058 KPC97718.1 JXJN01001399 MNUW01000075 OIO11218.1 CP018058 ASS99797.1 LDNZ01000185 KLR72735.1 KK571601 KFW07745.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000245037 UP000002320 UP000094527 UP000008820 UP000075884 UP000076408 UP000009192 UP000000304 UP000001292 UP000000803 UP000008792 UP000008711 UP000092553 UP000002282 UP000075840 UP000192221 UP000076858 UP000001070 UP000076753 UP000007819 UP000035314 UP000027227 UP000023566 UP000011251 UP000183832 UP000015500 UP000002223 UP000029337 UP000076542 UP000266922 UP000231952 UP000032522 UP000192799 UP000242188 UP000007801 UP000255043 UP000075883 UP000075424 UP000268350 UP000071670 UP000018339 UP000001819 UP000008744 UP000192362 UP000051885 UP000075517 UP000271374 UP000095300 UP000091820 UP000007266 UP000282892 UP000217162 UP000191301 UP000186030 UP000016424 UP000007161 UP000014211 UP000037948 UP000092443 UP000092460 UP000183732 UP000092445 UP000214728 UP000036182
UP000245037 UP000002320 UP000094527 UP000008820 UP000075884 UP000076408 UP000009192 UP000000304 UP000001292 UP000000803 UP000008792 UP000008711 UP000092553 UP000002282 UP000075840 UP000192221 UP000076858 UP000001070 UP000076753 UP000007819 UP000035314 UP000027227 UP000023566 UP000011251 UP000183832 UP000015500 UP000002223 UP000029337 UP000076542 UP000266922 UP000231952 UP000032522 UP000192799 UP000242188 UP000007801 UP000255043 UP000075883 UP000075424 UP000268350 UP000071670 UP000018339 UP000001819 UP000008744 UP000192362 UP000051885 UP000075517 UP000271374 UP000095300 UP000091820 UP000007266 UP000282892 UP000217162 UP000191301 UP000186030 UP000016424 UP000007161 UP000014211 UP000037948 UP000092443 UP000092460 UP000183732 UP000092445 UP000214728 UP000036182
Interpro
IPR011611
PfkB_dom
+ More
IPR029056 Ribokinase-like
IPR002173 Carboh/pur_kinase_PfkB_CS
IPR022830 Indigdn_synthA-like
IPR007342 PsuG
IPR036084 Ser_inhib-like_sf
IPR002919 TIL_dom
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR011991 ArsR-like_HTH
IPR012318 HTH_CRP
IPR002139 Ribo/fructo_kinase
IPR000485 AsnC-type_HTH_dom
IPR029056 Ribokinase-like
IPR002173 Carboh/pur_kinase_PfkB_CS
IPR022830 Indigdn_synthA-like
IPR007342 PsuG
IPR036084 Ser_inhib-like_sf
IPR002919 TIL_dom
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR011991 ArsR-like_HTH
IPR012318 HTH_CRP
IPR002139 Ribo/fructo_kinase
IPR000485 AsnC-type_HTH_dom
Gene 3D
CDD
ProteinModelPortal
H9JE34
A0A2H1WGX3
A0A2A4J5P7
A0A194R8M2
A0A194PQE0
A0A3S2NUB5
+ More
A0A212F6P9 A0A2P8YK20 A0A293LJT0 A0A293LIA6 B0WVP4 A0A1D2MGV0 Q17AQ9 A0A182NFW6 A0A182XXD0 B4KFG8 A0A0J9QU64 B4Q657 B4ICY9 Q9VPR3 Q8MYS6 B4LTN4 B3N7S1 A0A0M4E876 A0A0P5GTV1 A0A0P5LIS2 A0A0P5A5C8 A0A0P5LDJ7 A0A0P4ZBZ9 B4P2G1 A0A0P5JMQ7 A0A147BGQ2 A0A182I8S2 A0A1W4V2M9 A0A164XRJ0 B4JDR4 A0A167U815 A0A0P5WWE3 J9M3P9 A0A0P5W0Y9 A0A0N8ARS7 A0A0P5D2R0 A0A0P6GFA6 A0A0P6HQP4 A0A0G3XYQ0 A0A063YYC2 A0A023CLX3 L7ZQQ7 A0A1J1IFD5 A0A0P5A7V9 A0A0P6D064 S5YYB4 A0A0E0TAL8 A0A087LEC1 A0A161SHX7 A0A3L7CZ22 A0A2M9T249 A0A0D8BQU5 A0A1V9B3H9 A0A1B6D3E9 A0A1B6DRE2 A0A210PPV3 B3MLP7 A0A377FZG5 A0A182MJW6 A0A150M7Y9 A0A3B0JHG4 A0A142CZS0 V6VCM7 A0A2M4DS25 A0A0P5ZSC8 A0A2M4CRL9 Q29N57 B4G882 A0A1V9C1A7 A0A0Q0Y9E8 A0A150NF66 A0A3S0IXZ4 A0A3B0UKN6 A0A1I8PQD8 A0A1A9WN01 A0A139WH26 A0A3Q9QS05 A0A250DYF0 A0A2S2R297 A0A1L8E1E1 A0A1V4P819 A0A1Q5SR52 U2X8P4 H2J7F3 A0A069DYJ8 R9MSM4 A0A0N0Z1I2 A0A1A9XXR0 A0A1B0APG1 A0A1J4THC7 A0A1A9Z630 A0A226Q1I4 A0A0J0V357 A0A093IYC1
A0A212F6P9 A0A2P8YK20 A0A293LJT0 A0A293LIA6 B0WVP4 A0A1D2MGV0 Q17AQ9 A0A182NFW6 A0A182XXD0 B4KFG8 A0A0J9QU64 B4Q657 B4ICY9 Q9VPR3 Q8MYS6 B4LTN4 B3N7S1 A0A0M4E876 A0A0P5GTV1 A0A0P5LIS2 A0A0P5A5C8 A0A0P5LDJ7 A0A0P4ZBZ9 B4P2G1 A0A0P5JMQ7 A0A147BGQ2 A0A182I8S2 A0A1W4V2M9 A0A164XRJ0 B4JDR4 A0A167U815 A0A0P5WWE3 J9M3P9 A0A0P5W0Y9 A0A0N8ARS7 A0A0P5D2R0 A0A0P6GFA6 A0A0P6HQP4 A0A0G3XYQ0 A0A063YYC2 A0A023CLX3 L7ZQQ7 A0A1J1IFD5 A0A0P5A7V9 A0A0P6D064 S5YYB4 A0A0E0TAL8 A0A087LEC1 A0A161SHX7 A0A3L7CZ22 A0A2M9T249 A0A0D8BQU5 A0A1V9B3H9 A0A1B6D3E9 A0A1B6DRE2 A0A210PPV3 B3MLP7 A0A377FZG5 A0A182MJW6 A0A150M7Y9 A0A3B0JHG4 A0A142CZS0 V6VCM7 A0A2M4DS25 A0A0P5ZSC8 A0A2M4CRL9 Q29N57 B4G882 A0A1V9C1A7 A0A0Q0Y9E8 A0A150NF66 A0A3S0IXZ4 A0A3B0UKN6 A0A1I8PQD8 A0A1A9WN01 A0A139WH26 A0A3Q9QS05 A0A250DYF0 A0A2S2R297 A0A1L8E1E1 A0A1V4P819 A0A1Q5SR52 U2X8P4 H2J7F3 A0A069DYJ8 R9MSM4 A0A0N0Z1I2 A0A1A9XXR0 A0A1B0APG1 A0A1J4THC7 A0A1A9Z630 A0A226Q1I4 A0A0J0V357 A0A093IYC1
PDB
3KZH
E-value=8.49268e-11,
Score=154
Ontologies
GO
PANTHER
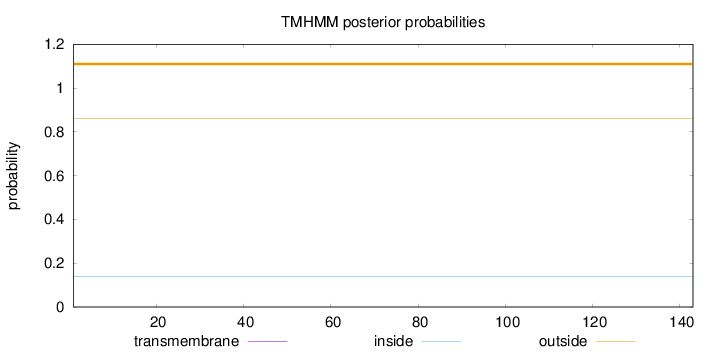
Topology
Length:
143
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00325
Exp number, first 60 AAs:
0.00201
Total prob of N-in:
0.14029
outside
1 - 143
Population Genetic Test Statistics
Pi
68.867422
Theta
75.51875
Tajima's D
0.256165
CLR
109.258111
CSRT
0.44142792860357
Interpretation
Uncertain
