Gene
KWMTBOMO08994
Pre Gene Modal
BGIBMGA007750
Annotation
PREDICTED:_uncharacterized_protein_LOC106107944_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.969
Sequence
CDS
ATGACTGCTGATAATGAATATCCTAAAGCATCAAATGCTACGCTCATTGGAGCCAGTATCACCTATGTGGGGGCTTTATTTCTCCTCCTTTCATTTGCTGGTCCTTACTGGATGGAATCATATCCAGAAATGTTCTCTCCATTCAAACATATGGGATTGTGGGAGTATTGCTTTGATAATTTCAGGTTCCCTGGTTACCGGTTTGACAAAAAATTTGATGGGTGTCACTATATCTACAGCCAAGAATATTATGCAATCAAGCATTGGCTGTTGCCTGGATGGTTAATGGCTGTGCAGTTTTTCGTGACAGTTGCGCTGATGTTGTCATTTGCAGCCCAGGTTTTATTGGCATGTGTCATTGTTCGTTGGCCATTGAAGACTGTGCTTCGTTATGAATGGATCTTTGTGTTCTATTCATTCATTATGGTAGCAGTTTCTAGTGCTTTCCTCTTTCTTGCTGTGGCAATATTCGGTGGGAACTGCTACAGACGTGACTGGCTCTTGTATCCTTCATTCAATGTTTTGTCTTGGTCCTATGCATTTGCGGTTATTGCCTTTATATTATTTGGTTTGGCATCGATTTTCCTGTATTTGGAGTTTCGAAAATTATATAAACTTCGCTTAGAGGCGAAAAATCTTGTCGCTCAAATGCAACATAGTCAACCTGAACACACCCTTAACCAACTCCGGGACCAATTGCAGCAACAGCAACAGCGTCAGTGGAAATGA
Protein
MTADNEYPKASNATLIGASITYVGALFLLLSFAGPYWMESYPEMFSPFKHMGLWEYCFDNFRFPGYRFDKKFDGCHYIYSQEYYAIKHWLLPGWLMAVQFFVTVALMLSFAAQVLLACVIVRWPLKTVLRYEWIFVFYSFIMVAVSSAFLFLAVAIFGGNCYRRDWLLYPSFNVLSWSYAFAVIAFILFGLASIFLYLEFRKLYKLRLEAKNLVAQMQHSQPEHTLNQLRDQLQQQQQRQWK
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JE03
A0A2H1WGF1
A0A437ASD9
A0A194PKU2
A0A194R7W0
A0A1E1WLE0
+ More
A0A212F6S1 A0A2A4J4C2 S4PEM3 A0A1Y9J0R6 A0A2C9GS04 A0A182UTB8 A0A182UCN9 A0A182LAN0 Q7QFW1 A0A182QIN4 A0A182S048 A0A182NFW5 W5JUR0 A0A2M4A9U5 A0A2M4A9U3 A0A084VC03 A0A2M3ZKL4 A0A182MWB8 A0A1W7R7Z0 A0A2M4BY76 A0A1L8DUW4 A0A1L8DUY3 A0A182P775 A0A0L7RIQ7 A0A182XXC9 A0A023EKX4 Q17AR0 A0A154P892 U5EDZ6 A0A182J5R3 A0A1J1IFD8 A0A2A3EE57 A0A088ARH1 A0A182F9F6 B0WVP5 A0A1Q3FPA1 A0A151ICK6 A0A0N0U6Z9 A0A067RM40 A0A1L8DUW9 A0A336LT38 A0A182JPE6 A0A026WZE6 A0A195FFB3 A0A182VXU0 J3JWU5 A0A195DY27 F4WZW1 A0A151XAG1 A0A158NGN2 A0A0K8TQ20 T1PJM1 A0A0C9PMU7 A0A1W4WKG3 D6WM28 B4MB24 A0A0A1WRU5 W8BGW6 B4JLQ9 E9ILA2 B4L752 A0A0T6B0F8 A0A1B0FAJ0 A0A1A9YQ85 A0A195B7T7 B4NQ02 Q29J10 B4GTK6 A0A1I8NXK9 A0A0J7L7F7 A0A232EFB3 K7J6J9 B3P9I8 A0A1B6JX14 B4PXQ2 B3LXH5 A0A3B0KME9 B4I9F0 E4NKM0 O76899 A0A0V0G5P8 A0A0A9WZF6 A0A1A9ULM5 A0A1B0A0A4 A0A069DR14 R4G3M3 A0A1B6FUQ3 A0A1W4VT65 A0A0K8UYT8 A0A2J7PUZ5 A0A034WBF9 A0A0C9PS97 E0VM69 A0A1Y1K7S1
A0A212F6S1 A0A2A4J4C2 S4PEM3 A0A1Y9J0R6 A0A2C9GS04 A0A182UTB8 A0A182UCN9 A0A182LAN0 Q7QFW1 A0A182QIN4 A0A182S048 A0A182NFW5 W5JUR0 A0A2M4A9U5 A0A2M4A9U3 A0A084VC03 A0A2M3ZKL4 A0A182MWB8 A0A1W7R7Z0 A0A2M4BY76 A0A1L8DUW4 A0A1L8DUY3 A0A182P775 A0A0L7RIQ7 A0A182XXC9 A0A023EKX4 Q17AR0 A0A154P892 U5EDZ6 A0A182J5R3 A0A1J1IFD8 A0A2A3EE57 A0A088ARH1 A0A182F9F6 B0WVP5 A0A1Q3FPA1 A0A151ICK6 A0A0N0U6Z9 A0A067RM40 A0A1L8DUW9 A0A336LT38 A0A182JPE6 A0A026WZE6 A0A195FFB3 A0A182VXU0 J3JWU5 A0A195DY27 F4WZW1 A0A151XAG1 A0A158NGN2 A0A0K8TQ20 T1PJM1 A0A0C9PMU7 A0A1W4WKG3 D6WM28 B4MB24 A0A0A1WRU5 W8BGW6 B4JLQ9 E9ILA2 B4L752 A0A0T6B0F8 A0A1B0FAJ0 A0A1A9YQ85 A0A195B7T7 B4NQ02 Q29J10 B4GTK6 A0A1I8NXK9 A0A0J7L7F7 A0A232EFB3 K7J6J9 B3P9I8 A0A1B6JX14 B4PXQ2 B3LXH5 A0A3B0KME9 B4I9F0 E4NKM0 O76899 A0A0V0G5P8 A0A0A9WZF6 A0A1A9ULM5 A0A1B0A0A4 A0A069DR14 R4G3M3 A0A1B6FUQ3 A0A1W4VT65 A0A0K8UYT8 A0A2J7PUZ5 A0A034WBF9 A0A0C9PS97 E0VM69 A0A1Y1K7S1
Pubmed
19121390
26354079
22118469
23622113
20966253
12364791
+ More
14747013 17210077 20920257 23761445 24438588 25244985 24945155 17510324 27004691 24845553 24508170 30249741 22516182 23537049 21719571 21347285 26369729 25315136 18362917 19820115 17994087 25830018 24495485 21282665 15632085 28648823 20075255 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26334808 25348373 20566863 28004739
14747013 17210077 20920257 23761445 24438588 25244985 24945155 17510324 27004691 24845553 24508170 30249741 22516182 23537049 21719571 21347285 26369729 25315136 18362917 19820115 17994087 25830018 24495485 21282665 15632085 28648823 20075255 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26334808 25348373 20566863 28004739
EMBL
BABH01002532
ODYU01008491
SOQ52141.1
RSAL01000996
RVE41000.1
KQ459602
+ More
KPI93349.1 KQ460615 KPJ13594.1 GDQN01003317 GDQN01003102 JAT87737.1 JAT87952.1 AGBW02009983 OWR49409.1 NWSH01003076 PCG66997.1 GAIX01007000 JAA85560.1 APCN01003108 AAAB01008844 EAA06100.5 AXCN02001148 ADMH02000438 ETN66489.1 GGFK01004179 MBW37500.1 GGFK01004191 MBW37512.1 ATLV01009963 KE524559 KFB35497.1 GGFM01008305 MBW29056.1 AXCM01001575 GEHC01000357 JAV47288.1 GGFJ01008821 MBW57962.1 GFDF01003874 JAV10210.1 GFDF01003836 JAV10248.1 KQ414584 KOC70621.1 GAPW01003771 GAPW01003770 JAC09827.1 BK008793 CH477331 DAA64998.1 EAT43326.1 KQ434826 KZC07428.1 GANO01004522 JAB55349.1 CVRI01000048 CRK98931.1 KZ288282 PBC29572.1 DS232130 EDS35652.1 GFDL01005727 JAV29318.1 KQ978053 KYM97769.1 KQ435726 KOX78232.1 KK852591 KDR20695.1 GFDF01003840 JAV10244.1 UFQT01000097 SSX19749.1 KK107063 QOIP01000005 EZA61193.1 RLU23078.1 KQ981625 KYN39051.1 APGK01051818 BT127713 KB741207 KB632327 AEE62675.1 ENN72890.1 ERL92505.1 KQ980107 KYN17617.1 GL888480 EGI60251.1 KQ982339 KYQ57352.1 ADTU01015151 GDAI01001139 JAI16464.1 KA648143 AFP62772.1 GBYB01002478 JAG72245.1 KQ971343 EFA04212.1 CH940655 EDW66433.1 GBXI01012720 JAD01572.1 GAMC01008583 JAB97972.1 CH916371 EDV91670.1 GL764074 EFZ18624.1 CH933812 EDW06198.1 LJIG01016418 KRT80660.1 CCAG010006943 CCAG010006944 KQ976574 KYM80310.1 CH964291 EDW86227.1 CH379063 EAL32492.2 CH479190 EDW25876.1 LBMM01000397 KMQ98461.1 NNAY01005071 OXU17041.1 AAZX01013268 CH954183 EDV45484.1 GECU01028266 GECU01019428 GECU01012449 GECU01003961 GECU01003383 JAS79440.1 JAS88278.1 JAS95257.1 JAT03746.1 JAT04324.1 CM000162 EDX00905.1 CH902617 EDV42819.1 OUUW01000011 SPP86291.1 CH480825 EDW43831.1 BT125817 ADR71726.1 AE014298 AL031027 AAF45632.1 AAG22385.2 CAA19840.1 GECL01002737 JAP03387.1 GBHO01029742 GBHO01029741 JAG13862.1 JAG13863.1 GBGD01002797 JAC86092.1 ACPB03020118 GAHY01001551 JAA75959.1 GECZ01015890 JAS53879.1 GDHF01020563 JAI31751.1 NEVH01021187 PNF20147.1 GAKP01006918 JAC52034.1 GBYB01004233 JAG74000.1 DS235291 EEB14475.1 GEZM01092461 JAV56551.1
KPI93349.1 KQ460615 KPJ13594.1 GDQN01003317 GDQN01003102 JAT87737.1 JAT87952.1 AGBW02009983 OWR49409.1 NWSH01003076 PCG66997.1 GAIX01007000 JAA85560.1 APCN01003108 AAAB01008844 EAA06100.5 AXCN02001148 ADMH02000438 ETN66489.1 GGFK01004179 MBW37500.1 GGFK01004191 MBW37512.1 ATLV01009963 KE524559 KFB35497.1 GGFM01008305 MBW29056.1 AXCM01001575 GEHC01000357 JAV47288.1 GGFJ01008821 MBW57962.1 GFDF01003874 JAV10210.1 GFDF01003836 JAV10248.1 KQ414584 KOC70621.1 GAPW01003771 GAPW01003770 JAC09827.1 BK008793 CH477331 DAA64998.1 EAT43326.1 KQ434826 KZC07428.1 GANO01004522 JAB55349.1 CVRI01000048 CRK98931.1 KZ288282 PBC29572.1 DS232130 EDS35652.1 GFDL01005727 JAV29318.1 KQ978053 KYM97769.1 KQ435726 KOX78232.1 KK852591 KDR20695.1 GFDF01003840 JAV10244.1 UFQT01000097 SSX19749.1 KK107063 QOIP01000005 EZA61193.1 RLU23078.1 KQ981625 KYN39051.1 APGK01051818 BT127713 KB741207 KB632327 AEE62675.1 ENN72890.1 ERL92505.1 KQ980107 KYN17617.1 GL888480 EGI60251.1 KQ982339 KYQ57352.1 ADTU01015151 GDAI01001139 JAI16464.1 KA648143 AFP62772.1 GBYB01002478 JAG72245.1 KQ971343 EFA04212.1 CH940655 EDW66433.1 GBXI01012720 JAD01572.1 GAMC01008583 JAB97972.1 CH916371 EDV91670.1 GL764074 EFZ18624.1 CH933812 EDW06198.1 LJIG01016418 KRT80660.1 CCAG010006943 CCAG010006944 KQ976574 KYM80310.1 CH964291 EDW86227.1 CH379063 EAL32492.2 CH479190 EDW25876.1 LBMM01000397 KMQ98461.1 NNAY01005071 OXU17041.1 AAZX01013268 CH954183 EDV45484.1 GECU01028266 GECU01019428 GECU01012449 GECU01003961 GECU01003383 JAS79440.1 JAS88278.1 JAS95257.1 JAT03746.1 JAT04324.1 CM000162 EDX00905.1 CH902617 EDV42819.1 OUUW01000011 SPP86291.1 CH480825 EDW43831.1 BT125817 ADR71726.1 AE014298 AL031027 AAF45632.1 AAG22385.2 CAA19840.1 GECL01002737 JAP03387.1 GBHO01029742 GBHO01029741 JAG13862.1 JAG13863.1 GBGD01002797 JAC86092.1 ACPB03020118 GAHY01001551 JAA75959.1 GECZ01015890 JAS53879.1 GDHF01020563 JAI31751.1 NEVH01021187 PNF20147.1 GAKP01006918 JAC52034.1 GBYB01004233 JAG74000.1 DS235291 EEB14475.1 GEZM01092461 JAV56551.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000218220
+ More
UP000076407 UP000075840 UP000075903 UP000075902 UP000075882 UP000007062 UP000075886 UP000075900 UP000075884 UP000000673 UP000030765 UP000075883 UP000075885 UP000053825 UP000076408 UP000008820 UP000076502 UP000075880 UP000183832 UP000242457 UP000005203 UP000069272 UP000002320 UP000078542 UP000053105 UP000027135 UP000075881 UP000053097 UP000279307 UP000078541 UP000075920 UP000019118 UP000030742 UP000078492 UP000007755 UP000075809 UP000005205 UP000095301 UP000192223 UP000007266 UP000008792 UP000001070 UP000009192 UP000092444 UP000092443 UP000078540 UP000007798 UP000001819 UP000008744 UP000095300 UP000036403 UP000215335 UP000002358 UP000008711 UP000002282 UP000007801 UP000268350 UP000001292 UP000000803 UP000078200 UP000092445 UP000015103 UP000192221 UP000235965 UP000009046
UP000076407 UP000075840 UP000075903 UP000075902 UP000075882 UP000007062 UP000075886 UP000075900 UP000075884 UP000000673 UP000030765 UP000075883 UP000075885 UP000053825 UP000076408 UP000008820 UP000076502 UP000075880 UP000183832 UP000242457 UP000005203 UP000069272 UP000002320 UP000078542 UP000053105 UP000027135 UP000075881 UP000053097 UP000279307 UP000078541 UP000075920 UP000019118 UP000030742 UP000078492 UP000007755 UP000075809 UP000005205 UP000095301 UP000192223 UP000007266 UP000008792 UP000001070 UP000009192 UP000092444 UP000092443 UP000078540 UP000007798 UP000001819 UP000008744 UP000095300 UP000036403 UP000215335 UP000002358 UP000008711 UP000002282 UP000007801 UP000268350 UP000001292 UP000000803 UP000078200 UP000092445 UP000015103 UP000192221 UP000235965 UP000009046
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9JE03
A0A2H1WGF1
A0A437ASD9
A0A194PKU2
A0A194R7W0
A0A1E1WLE0
+ More
A0A212F6S1 A0A2A4J4C2 S4PEM3 A0A1Y9J0R6 A0A2C9GS04 A0A182UTB8 A0A182UCN9 A0A182LAN0 Q7QFW1 A0A182QIN4 A0A182S048 A0A182NFW5 W5JUR0 A0A2M4A9U5 A0A2M4A9U3 A0A084VC03 A0A2M3ZKL4 A0A182MWB8 A0A1W7R7Z0 A0A2M4BY76 A0A1L8DUW4 A0A1L8DUY3 A0A182P775 A0A0L7RIQ7 A0A182XXC9 A0A023EKX4 Q17AR0 A0A154P892 U5EDZ6 A0A182J5R3 A0A1J1IFD8 A0A2A3EE57 A0A088ARH1 A0A182F9F6 B0WVP5 A0A1Q3FPA1 A0A151ICK6 A0A0N0U6Z9 A0A067RM40 A0A1L8DUW9 A0A336LT38 A0A182JPE6 A0A026WZE6 A0A195FFB3 A0A182VXU0 J3JWU5 A0A195DY27 F4WZW1 A0A151XAG1 A0A158NGN2 A0A0K8TQ20 T1PJM1 A0A0C9PMU7 A0A1W4WKG3 D6WM28 B4MB24 A0A0A1WRU5 W8BGW6 B4JLQ9 E9ILA2 B4L752 A0A0T6B0F8 A0A1B0FAJ0 A0A1A9YQ85 A0A195B7T7 B4NQ02 Q29J10 B4GTK6 A0A1I8NXK9 A0A0J7L7F7 A0A232EFB3 K7J6J9 B3P9I8 A0A1B6JX14 B4PXQ2 B3LXH5 A0A3B0KME9 B4I9F0 E4NKM0 O76899 A0A0V0G5P8 A0A0A9WZF6 A0A1A9ULM5 A0A1B0A0A4 A0A069DR14 R4G3M3 A0A1B6FUQ3 A0A1W4VT65 A0A0K8UYT8 A0A2J7PUZ5 A0A034WBF9 A0A0C9PS97 E0VM69 A0A1Y1K7S1
A0A212F6S1 A0A2A4J4C2 S4PEM3 A0A1Y9J0R6 A0A2C9GS04 A0A182UTB8 A0A182UCN9 A0A182LAN0 Q7QFW1 A0A182QIN4 A0A182S048 A0A182NFW5 W5JUR0 A0A2M4A9U5 A0A2M4A9U3 A0A084VC03 A0A2M3ZKL4 A0A182MWB8 A0A1W7R7Z0 A0A2M4BY76 A0A1L8DUW4 A0A1L8DUY3 A0A182P775 A0A0L7RIQ7 A0A182XXC9 A0A023EKX4 Q17AR0 A0A154P892 U5EDZ6 A0A182J5R3 A0A1J1IFD8 A0A2A3EE57 A0A088ARH1 A0A182F9F6 B0WVP5 A0A1Q3FPA1 A0A151ICK6 A0A0N0U6Z9 A0A067RM40 A0A1L8DUW9 A0A336LT38 A0A182JPE6 A0A026WZE6 A0A195FFB3 A0A182VXU0 J3JWU5 A0A195DY27 F4WZW1 A0A151XAG1 A0A158NGN2 A0A0K8TQ20 T1PJM1 A0A0C9PMU7 A0A1W4WKG3 D6WM28 B4MB24 A0A0A1WRU5 W8BGW6 B4JLQ9 E9ILA2 B4L752 A0A0T6B0F8 A0A1B0FAJ0 A0A1A9YQ85 A0A195B7T7 B4NQ02 Q29J10 B4GTK6 A0A1I8NXK9 A0A0J7L7F7 A0A232EFB3 K7J6J9 B3P9I8 A0A1B6JX14 B4PXQ2 B3LXH5 A0A3B0KME9 B4I9F0 E4NKM0 O76899 A0A0V0G5P8 A0A0A9WZF6 A0A1A9ULM5 A0A1B0A0A4 A0A069DR14 R4G3M3 A0A1B6FUQ3 A0A1W4VT65 A0A0K8UYT8 A0A2J7PUZ5 A0A034WBF9 A0A0C9PS97 E0VM69 A0A1Y1K7S1
Ontologies
GO
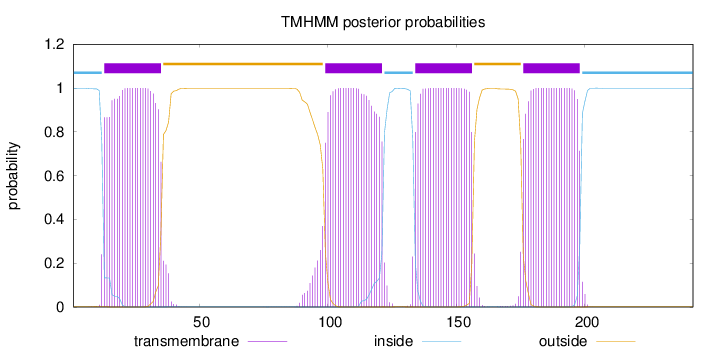
Topology
Length:
242
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
92.17056
Exp number, first 60 AAs:
22.71964
Total prob of N-in:
0.99851
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 98
TMhelix
99 - 121
inside
122 - 133
TMhelix
134 - 156
outside
157 - 175
TMhelix
176 - 198
inside
199 - 242
Population Genetic Test Statistics
Pi
255.125539
Theta
202.583997
Tajima's D
1.158777
CLR
0
CSRT
0.697165141742913
Interpretation
Uncertain
