Gene
KWMTBOMO08993
Pre Gene Modal
BGIBMGA007749
Annotation
PREDICTED:_uncharacterized_protein_LOC106138409_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.977
Sequence
CDS
ATGAAGAATAGAAGTTTGGCCGGTAATTGTGGCATTGGCGTTTTTGTTATTGCCCTTGTCACAGTCGCCTTGGCTTTCGGCACACCGAGTTGGTTGGTTAGCGACTCTAGAATAAGGAGTGCAAGATTGGACAGGCTTGGATTATGGAACTACTGCTTCAGATCACTGCCAGATCCATTAGATCAGCACCAGAGACGGTTTTTCGTGGGTTGTCGTTGGGTCTATGACCCGTTTACTACAGGGTATGATAAGATTCGAGGCTATTTGTTACCAGGATTTATGATAGCAACTCAATTCTTCTTCACACTGTGCCTCCTCGGAGTGTTAATAAGTACCATATTGGTATTGATATTCTTTCTTTGCTGTGGACCCGACCAAAGCAGATTCATCTTATTGATCAAAAGCATTGGATTCATTATGTTGGGAGCTGGAATAAGTGGTGCAATTGCTGTTATAGTATTTGCATGTTTGGGTAACACCGATGGGTGGATGCCAGATCATCCCAACAACTATTTAGGATGGTCATTCGGATTGGGCGTGGTCGGCGCTATAGCGTGTATTGTTGCTTCCGCTTTGTTTTTGACCGAAGCTAATATCCAACGTAAGAAACGCAATAAACTAAAAGAGTCACAAGCGCGCTTCGAAATGGAATACGAATCAAAGGCTTAA
Protein
MKNRSLAGNCGIGVFVIALVTVALAFGTPSWLVSDSRIRSARLDRLGLWNYCFRSLPDPLDQHQRRFFVGCRWVYDPFTTGYDKIRGYLLPGFMIATQFFFTLCLLGVLISTILVLIFFLCCGPDQSRFILLIKSIGFIMLGAGISGAIAVIVFACLGNTDGWMPDHPNNYLGWSFGLGVVGAIACIVASALFLTEANIQRKKRNKLKESQARFEMEYESKA
Summary
Uniprot
H9JE02
A0A437ASB9
A0A2H1WGD5
G3LHK3
A0A2A4J5H7
A0A194R700
+ More
A0A212F6R0 Q17AR2 A0A1Y9J0T1 A0A1Y9J049 A0A2C9GS06 A0A182V5K4 A0A182TU61 F5HJC0 Q17AR1 A0A182R5Y0 A0A453YN79 A0A182LAN3 A0A182NFW3 A0A182ING8 A0A182MTS8 A0A1Q3FNL4 A0A1Q3FN83 A0A2M4AMG1 A0A2M3ZEE4 A0A2J7PUY6 W5JU69 B0WVP6 A0A2M4BZ64 A0A067RJK2 A0A336LNX0 A0A336LS12 A0A182JPE6 D6WM27 A0A182HDW3 A0A023EK40 A0A0K8VYY7 A0A034V8K6 A0A0A1WV27 A0A023ELM6 B4HUG2 A0A1W4W4L0 B4QRL2 Q9VRM3 A0A1L8D9I2 A0A1L8D8Q7 B3NGC0 A0A2P8YK17 B4PIU3 A0A336LPC2 A0A1B6HDI5 A0A1B6ERT9 A0A0K8TR99 A0A026X1D9 A0A0L0C716 A0A182XXC8 A0A151XAG2 A0A023ENH6 A0A1B6KLF5 T1PE53 A0A1I8MJ06 A0A182P775 A0A0L7LDI7 W8B1E9 B3M3P8 A0A0M3QVX1 A0A158NGN3 E2BAR2 A0A151ICV5 A0A0L7RI04 F4WZW0 A0A0P4VPD6 T1HMN7 A0A224XJG0 A0A0V0G807 A0A1W4WJ90 U4UEV2 N6SYN3 J3JU96 A0A0N0BIT9 A0A195DXQ9 A0A195FEE4 A0A023F3L5 Q2LZM4 B4H3C9 A0A1A9Y9N4 A0A1Y1LGC3 B4MLT3 B4LBD8 K7J6K0 Q8T047 A0A3B0JW35 B4KVB2 A0A1I8NTW1 U5EWM7 B4IXS2 A0A1A9X2X0 A0A232EFE4 A0A069DQV7 A0A0A9XV34
A0A212F6R0 Q17AR2 A0A1Y9J0T1 A0A1Y9J049 A0A2C9GS06 A0A182V5K4 A0A182TU61 F5HJC0 Q17AR1 A0A182R5Y0 A0A453YN79 A0A182LAN3 A0A182NFW3 A0A182ING8 A0A182MTS8 A0A1Q3FNL4 A0A1Q3FN83 A0A2M4AMG1 A0A2M3ZEE4 A0A2J7PUY6 W5JU69 B0WVP6 A0A2M4BZ64 A0A067RJK2 A0A336LNX0 A0A336LS12 A0A182JPE6 D6WM27 A0A182HDW3 A0A023EK40 A0A0K8VYY7 A0A034V8K6 A0A0A1WV27 A0A023ELM6 B4HUG2 A0A1W4W4L0 B4QRL2 Q9VRM3 A0A1L8D9I2 A0A1L8D8Q7 B3NGC0 A0A2P8YK17 B4PIU3 A0A336LPC2 A0A1B6HDI5 A0A1B6ERT9 A0A0K8TR99 A0A026X1D9 A0A0L0C716 A0A182XXC8 A0A151XAG2 A0A023ENH6 A0A1B6KLF5 T1PE53 A0A1I8MJ06 A0A182P775 A0A0L7LDI7 W8B1E9 B3M3P8 A0A0M3QVX1 A0A158NGN3 E2BAR2 A0A151ICV5 A0A0L7RI04 F4WZW0 A0A0P4VPD6 T1HMN7 A0A224XJG0 A0A0V0G807 A0A1W4WJ90 U4UEV2 N6SYN3 J3JU96 A0A0N0BIT9 A0A195DXQ9 A0A195FEE4 A0A023F3L5 Q2LZM4 B4H3C9 A0A1A9Y9N4 A0A1Y1LGC3 B4MLT3 B4LBD8 K7J6K0 Q8T047 A0A3B0JW35 B4KVB2 A0A1I8NTW1 U5EWM7 B4IXS2 A0A1A9X2X0 A0A232EFE4 A0A069DQV7 A0A0A9XV34
Pubmed
19121390
21840855
26354079
22118469
17510324
12364791
+ More
14747013 17210077 27004691 20966253 20920257 23761445 24845553 18362917 19820115 26483478 24945155 25348373 25830018 17994087 22936249 10731132 12537568 12537572 12537573 12537574 14734539 16110336 17569856 17569867 26109357 26109356 29403074 17550304 26369729 24508170 30249741 26108605 25244985 25315136 26227816 24495485 21347285 20798317 21719571 27129103 23537049 22516182 25474469 15632085 28004739 18057021 20075255 28648823 26334808 25401762
14747013 17210077 27004691 20966253 20920257 23761445 24845553 18362917 19820115 26483478 24945155 25348373 25830018 17994087 22936249 10731132 12537568 12537572 12537573 12537574 14734539 16110336 17569856 17569867 26109357 26109356 29403074 17550304 26369729 24508170 30249741 26108605 25244985 25315136 26227816 24495485 21347285 20798317 21719571 27129103 23537049 22516182 25474469 15632085 28004739 18057021 20075255 28648823 26334808 25401762
EMBL
BABH01002532
RSAL01000996
RVE40999.1
ODYU01008491
SOQ52140.1
JN383806
+ More
AEN04482.1 NWSH01003076 PCG66996.1 KQ460615 KPJ13593.1 AGBW02009983 OWR49408.1 CH477331 EAT43323.1 APCN01003108 AAAB01008844 EAA05964.4 EGK96380.1 EGK96381.1 BK008794 DAA64999.1 EAT43324.1 EAT43325.1 AXCM01001575 GFDL01005972 JAV29073.1 GFDL01006024 JAV29021.1 GGFK01008643 MBW41964.1 GGFM01006114 MBW26865.1 NEVH01021187 PNF20148.1 ADMH02000438 ETN66490.1 DS232130 EDS35653.1 GGFJ01009153 MBW58294.1 KK852591 KDR20696.1 UFQT01000097 SSX19752.1 SSX19751.1 KQ971343 EFA04211.1 JXUM01035861 JXUM01035862 JXUM01062254 JXUM01062255 KQ562191 KQ561076 KXJ76453.1 KXJ79792.1 GAPW01004121 GAPW01004120 JAC09477.1 GDHF01008227 JAI44087.1 GAKP01020108 JAC38844.1 GBXI01011937 JAD02355.1 GAPW01003447 JAC10151.1 CH480817 EDW50583.1 CM000363 CM002912 EDX09323.1 KMY97751.1 AE014296 AY423544 AAF50772.3 AAR24293.1 GFDF01010971 JAV03113.1 GFDF01011235 JAV02849.1 CH954178 EDV51021.1 PYGN01000538 PSN44601.1 CM000159 EDW93513.1 SSX19750.1 GECU01035011 JAS72695.1 GECZ01029181 GECZ01007214 JAS40588.1 JAS62555.1 GDAI01001153 JAI16450.1 KK107063 QOIP01000005 EZA61194.1 RLU23077.1 JRES01000810 KNC28198.1 KQ982339 KYQ57353.1 GAPW01003634 JAC09964.1 GEBQ01027706 JAT12271.1 KA646233 AFP60862.1 JTDY01001580 KOB73465.1 GAMC01015712 JAB90843.1 CH902618 EDV40341.1 CP012525 ALC43143.1 ADTU01015149 GL446831 EFN87197.1 KQ978053 KYM97768.1 KQ414584 KOC70622.1 GL888480 EGI60250.1 GDKW01002250 JAI54345.1 ACPB03015130 GFTR01003809 JAW12617.1 GECL01002069 JAP04055.1 KB632327 ERL92504.1 APGK01051818 KB741207 ENN72889.1 BT126809 AEE61771.1 KQ435726 KOX78231.1 KQ980107 KYN17616.1 KQ981625 KYN39050.1 GBBI01002756 JAC15956.1 CH379069 EAL29483.2 CH479206 EDW30869.1 GEZM01059687 JAV71400.1 CH963847 EDW73009.2 CH940647 EDW69726.2 KRF84540.1 AAZX01013268 AY069562 AAL39707.1 OUUW01000002 SPP76931.1 CH933809 EDW18355.1 KRG06144.1 GANO01001346 JAB58525.1 CH916366 EDV96443.1 NNAY01005071 OXU17042.1 GBGD01002798 JAC86091.1 GBHO01019810 JAG23794.1
AEN04482.1 NWSH01003076 PCG66996.1 KQ460615 KPJ13593.1 AGBW02009983 OWR49408.1 CH477331 EAT43323.1 APCN01003108 AAAB01008844 EAA05964.4 EGK96380.1 EGK96381.1 BK008794 DAA64999.1 EAT43324.1 EAT43325.1 AXCM01001575 GFDL01005972 JAV29073.1 GFDL01006024 JAV29021.1 GGFK01008643 MBW41964.1 GGFM01006114 MBW26865.1 NEVH01021187 PNF20148.1 ADMH02000438 ETN66490.1 DS232130 EDS35653.1 GGFJ01009153 MBW58294.1 KK852591 KDR20696.1 UFQT01000097 SSX19752.1 SSX19751.1 KQ971343 EFA04211.1 JXUM01035861 JXUM01035862 JXUM01062254 JXUM01062255 KQ562191 KQ561076 KXJ76453.1 KXJ79792.1 GAPW01004121 GAPW01004120 JAC09477.1 GDHF01008227 JAI44087.1 GAKP01020108 JAC38844.1 GBXI01011937 JAD02355.1 GAPW01003447 JAC10151.1 CH480817 EDW50583.1 CM000363 CM002912 EDX09323.1 KMY97751.1 AE014296 AY423544 AAF50772.3 AAR24293.1 GFDF01010971 JAV03113.1 GFDF01011235 JAV02849.1 CH954178 EDV51021.1 PYGN01000538 PSN44601.1 CM000159 EDW93513.1 SSX19750.1 GECU01035011 JAS72695.1 GECZ01029181 GECZ01007214 JAS40588.1 JAS62555.1 GDAI01001153 JAI16450.1 KK107063 QOIP01000005 EZA61194.1 RLU23077.1 JRES01000810 KNC28198.1 KQ982339 KYQ57353.1 GAPW01003634 JAC09964.1 GEBQ01027706 JAT12271.1 KA646233 AFP60862.1 JTDY01001580 KOB73465.1 GAMC01015712 JAB90843.1 CH902618 EDV40341.1 CP012525 ALC43143.1 ADTU01015149 GL446831 EFN87197.1 KQ978053 KYM97768.1 KQ414584 KOC70622.1 GL888480 EGI60250.1 GDKW01002250 JAI54345.1 ACPB03015130 GFTR01003809 JAW12617.1 GECL01002069 JAP04055.1 KB632327 ERL92504.1 APGK01051818 KB741207 ENN72889.1 BT126809 AEE61771.1 KQ435726 KOX78231.1 KQ980107 KYN17616.1 KQ981625 KYN39050.1 GBBI01002756 JAC15956.1 CH379069 EAL29483.2 CH479206 EDW30869.1 GEZM01059687 JAV71400.1 CH963847 EDW73009.2 CH940647 EDW69726.2 KRF84540.1 AAZX01013268 AY069562 AAL39707.1 OUUW01000002 SPP76931.1 CH933809 EDW18355.1 KRG06144.1 GANO01001346 JAB58525.1 CH916366 EDV96443.1 NNAY01005071 OXU17042.1 GBGD01002798 JAC86091.1 GBHO01019810 JAG23794.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000007151
UP000008820
+ More
UP000076407 UP000075840 UP000075903 UP000075902 UP000007062 UP000075900 UP000075882 UP000075884 UP000075880 UP000075883 UP000235965 UP000000673 UP000002320 UP000027135 UP000075881 UP000007266 UP000069940 UP000249989 UP000001292 UP000192221 UP000000304 UP000000803 UP000008711 UP000245037 UP000002282 UP000053097 UP000279307 UP000037069 UP000076408 UP000075809 UP000095301 UP000075885 UP000037510 UP000007801 UP000092553 UP000005205 UP000008237 UP000078542 UP000053825 UP000007755 UP000015103 UP000192223 UP000030742 UP000019118 UP000053105 UP000078492 UP000078541 UP000001819 UP000008744 UP000092443 UP000007798 UP000008792 UP000002358 UP000268350 UP000009192 UP000095300 UP000001070 UP000091820 UP000215335
UP000076407 UP000075840 UP000075903 UP000075902 UP000007062 UP000075900 UP000075882 UP000075884 UP000075880 UP000075883 UP000235965 UP000000673 UP000002320 UP000027135 UP000075881 UP000007266 UP000069940 UP000249989 UP000001292 UP000192221 UP000000304 UP000000803 UP000008711 UP000245037 UP000002282 UP000053097 UP000279307 UP000037069 UP000076408 UP000075809 UP000095301 UP000075885 UP000037510 UP000007801 UP000092553 UP000005205 UP000008237 UP000078542 UP000053825 UP000007755 UP000015103 UP000192223 UP000030742 UP000019118 UP000053105 UP000078492 UP000078541 UP000001819 UP000008744 UP000092443 UP000007798 UP000008792 UP000002358 UP000268350 UP000009192 UP000095300 UP000001070 UP000091820 UP000215335
Pfam
PF13903 Claudin_2
SUPFAM
SSF103473
SSF103473
ProteinModelPortal
H9JE02
A0A437ASB9
A0A2H1WGD5
G3LHK3
A0A2A4J5H7
A0A194R700
+ More
A0A212F6R0 Q17AR2 A0A1Y9J0T1 A0A1Y9J049 A0A2C9GS06 A0A182V5K4 A0A182TU61 F5HJC0 Q17AR1 A0A182R5Y0 A0A453YN79 A0A182LAN3 A0A182NFW3 A0A182ING8 A0A182MTS8 A0A1Q3FNL4 A0A1Q3FN83 A0A2M4AMG1 A0A2M3ZEE4 A0A2J7PUY6 W5JU69 B0WVP6 A0A2M4BZ64 A0A067RJK2 A0A336LNX0 A0A336LS12 A0A182JPE6 D6WM27 A0A182HDW3 A0A023EK40 A0A0K8VYY7 A0A034V8K6 A0A0A1WV27 A0A023ELM6 B4HUG2 A0A1W4W4L0 B4QRL2 Q9VRM3 A0A1L8D9I2 A0A1L8D8Q7 B3NGC0 A0A2P8YK17 B4PIU3 A0A336LPC2 A0A1B6HDI5 A0A1B6ERT9 A0A0K8TR99 A0A026X1D9 A0A0L0C716 A0A182XXC8 A0A151XAG2 A0A023ENH6 A0A1B6KLF5 T1PE53 A0A1I8MJ06 A0A182P775 A0A0L7LDI7 W8B1E9 B3M3P8 A0A0M3QVX1 A0A158NGN3 E2BAR2 A0A151ICV5 A0A0L7RI04 F4WZW0 A0A0P4VPD6 T1HMN7 A0A224XJG0 A0A0V0G807 A0A1W4WJ90 U4UEV2 N6SYN3 J3JU96 A0A0N0BIT9 A0A195DXQ9 A0A195FEE4 A0A023F3L5 Q2LZM4 B4H3C9 A0A1A9Y9N4 A0A1Y1LGC3 B4MLT3 B4LBD8 K7J6K0 Q8T047 A0A3B0JW35 B4KVB2 A0A1I8NTW1 U5EWM7 B4IXS2 A0A1A9X2X0 A0A232EFE4 A0A069DQV7 A0A0A9XV34
A0A212F6R0 Q17AR2 A0A1Y9J0T1 A0A1Y9J049 A0A2C9GS06 A0A182V5K4 A0A182TU61 F5HJC0 Q17AR1 A0A182R5Y0 A0A453YN79 A0A182LAN3 A0A182NFW3 A0A182ING8 A0A182MTS8 A0A1Q3FNL4 A0A1Q3FN83 A0A2M4AMG1 A0A2M3ZEE4 A0A2J7PUY6 W5JU69 B0WVP6 A0A2M4BZ64 A0A067RJK2 A0A336LNX0 A0A336LS12 A0A182JPE6 D6WM27 A0A182HDW3 A0A023EK40 A0A0K8VYY7 A0A034V8K6 A0A0A1WV27 A0A023ELM6 B4HUG2 A0A1W4W4L0 B4QRL2 Q9VRM3 A0A1L8D9I2 A0A1L8D8Q7 B3NGC0 A0A2P8YK17 B4PIU3 A0A336LPC2 A0A1B6HDI5 A0A1B6ERT9 A0A0K8TR99 A0A026X1D9 A0A0L0C716 A0A182XXC8 A0A151XAG2 A0A023ENH6 A0A1B6KLF5 T1PE53 A0A1I8MJ06 A0A182P775 A0A0L7LDI7 W8B1E9 B3M3P8 A0A0M3QVX1 A0A158NGN3 E2BAR2 A0A151ICV5 A0A0L7RI04 F4WZW0 A0A0P4VPD6 T1HMN7 A0A224XJG0 A0A0V0G807 A0A1W4WJ90 U4UEV2 N6SYN3 J3JU96 A0A0N0BIT9 A0A195DXQ9 A0A195FEE4 A0A023F3L5 Q2LZM4 B4H3C9 A0A1A9Y9N4 A0A1Y1LGC3 B4MLT3 B4LBD8 K7J6K0 Q8T047 A0A3B0JW35 B4KVB2 A0A1I8NTW1 U5EWM7 B4IXS2 A0A1A9X2X0 A0A232EFE4 A0A069DQV7 A0A0A9XV34
Ontologies
GO
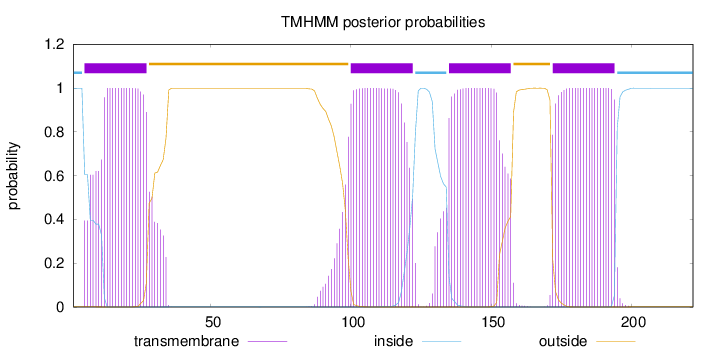
Topology
Length:
222
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
93.54755
Exp number, first 60 AAs:
22.42556
Total prob of N-in:
0.99915
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 99
TMhelix
100 - 122
inside
123 - 134
TMhelix
135 - 157
outside
158 - 171
TMhelix
172 - 194
inside
195 - 222
Population Genetic Test Statistics
Pi
204.345861
Theta
170.162477
Tajima's D
0.609863
CLR
0.599049
CSRT
0.549222538873056
Interpretation
Uncertain
