Gene
KWMTBOMO08992
Pre Gene Modal
BGIBMGA007748
Annotation
Kune-kune_[Operophtera_brumata]
Location in the cell
PlasmaMembrane Reliability : 4.901
Sequence
CDS
ATGACGAAATCTAAGACAGGCTACTTTGCTTTGGGATTTTTCGCATTCGCAGCTCTTTTTATTATAATAGCATTTGGAAGTCCATATTGGTTGGTAACTGATGGGAAGCTAAAGGATTCAAAGTTTCTGAAAATAGGTTTGTGGGAAGTATGTTTCAACAAATTCGAAGACGTTCATCACTGGTACGATACCGTCTTTAACAGCTGCTGGTGGATATTTGAAGAAGAATTCTATATTATACATGATATTTTATTGCCTGGTTTTTTCATTGCGACACAGTTCTTCTTCACCATAACATTGTGTTGTGTTCTGCTTTCGATGTTTTTGGTTTTTCTTTATTTAAAGAAGGACAAGGATGATGATGACTATTTGACACTATTGGTAACTTTAGGAACTGTGTTAGTGATCGGAGGCTTTTCTGGAATCATATCAGTTGTGACATTTGGAGCCAGAGGTGATGGGCGTGATTGGATGCCCCATTGGCAGAATAATGACTTGGGTTGGGCTTATGCTCTTGGAGTTGTGGGTGTAATATCACTGTTTCCTGCTGGTATACTGTTTTTAATTGTAGCCAGAGTACACAAATACAAAAAACTGCATGAGATGCAGAGTCGTGAGCCATCCGCTTACAATATGCATGAAAGGAACCTTGGTTATACCAGAGGACACACCGATATTTAA
Protein
MTKSKTGYFALGFFAFAALFIIIAFGSPYWLVTDGKLKDSKFLKIGLWEVCFNKFEDVHHWYDTVFNSCWWIFEEEFYIIHDILLPGFFIATQFFFTITLCCVLLSMFLVFLYLKKDKDDDDYLTLLVTLGTVLVIGGFSGIISVVTFGARGDGRDWMPHWQNNDLGWAYALGVVGVISLFPAGILFLIVARVHKYKKLHEMQSREPSAYNMHERNLGYTRGHTDI
Summary
Uniprot
H9JE01
A0A2H1WGC9
A0A2A4IVM1
A0A0L7LE33
I4DML7
A0A212F6P1
+ More
A0A194RCF9 A0A437B2Q1 S4PMX5 A0A194PIU8 A0A1L8D9V5 Q17AR3 A0A023EK76 B4LPH4 A0A182VXT9 B4NZC2 Q293G9 A0A023EMI3 T1P9U2 A0A182MVR1 B4II00 W5JQV5 A0A182R5Y2 A0A0J9R6A3 A0A182JPE8 B4GBI8 A0A182P773 A0A182VBD2 A0A0L0C5L0 Q8SY89 A1Z6H0 A0A182LAN6 A0A182TTI9 A0A182WV45 Q7PS72 A0A182I8R9 A0A1I8PZ66 B3N3J4 A0A2M3Z6Y5 A0A2M4BYT4 A0A1A9X4R2 A0A2M4AI51 A0A2M4AGN5 B4KLR9 A0A182Q7Z8 W8C0J3 A0A0M4E413 A0A067RA88 A0A2J7PUY9 B4MJ02 B0WVP8 A0A1Q3FJX5 A0A1B0A3P9 A0A1A9V241 A0A0K8UIY8 A0A1A9XQS1 A0A1B0B5C3 U5EFK3 A0A034W5J7 A0A0A1XC51 A0A182J731 B4J9F3 B3MJU1 A0A1W4W7V9 A0A2P8YK55 A0A182NFW1 A0A182XXC8 D6WM26 A0A1W4W956 A0A026WYS3 A0A0J7L7A3 J3JUI6 A0A2S2R3I0 A0A0C9RMG8 A0A224XJM7 A0A0K8TRD6 A0A182F9F6 A0A1J1IKF5 A0A151ICF6 A0A023F2K1 A0A151XAG3 F4WZV9 A0A158NGN4 A0A2A3EDL4 A0A088ARG9 T1HN22 A0A154P662 J9K1B7 A0A0M9A6H3 E2BAR1 A0A1Y1N351 A0A1Y1N4C0 A0A1B6I177 A0A0L7RIJ5 E0VZL1 A0A1B6CZL8 A0A0A9Z8Z2 A0A1B6GJJ7 A0A1L8D7K1 K7J6K1
A0A194RCF9 A0A437B2Q1 S4PMX5 A0A194PIU8 A0A1L8D9V5 Q17AR3 A0A023EK76 B4LPH4 A0A182VXT9 B4NZC2 Q293G9 A0A023EMI3 T1P9U2 A0A182MVR1 B4II00 W5JQV5 A0A182R5Y2 A0A0J9R6A3 A0A182JPE8 B4GBI8 A0A182P773 A0A182VBD2 A0A0L0C5L0 Q8SY89 A1Z6H0 A0A182LAN6 A0A182TTI9 A0A182WV45 Q7PS72 A0A182I8R9 A0A1I8PZ66 B3N3J4 A0A2M3Z6Y5 A0A2M4BYT4 A0A1A9X4R2 A0A2M4AI51 A0A2M4AGN5 B4KLR9 A0A182Q7Z8 W8C0J3 A0A0M4E413 A0A067RA88 A0A2J7PUY9 B4MJ02 B0WVP8 A0A1Q3FJX5 A0A1B0A3P9 A0A1A9V241 A0A0K8UIY8 A0A1A9XQS1 A0A1B0B5C3 U5EFK3 A0A034W5J7 A0A0A1XC51 A0A182J731 B4J9F3 B3MJU1 A0A1W4W7V9 A0A2P8YK55 A0A182NFW1 A0A182XXC8 D6WM26 A0A1W4W956 A0A026WYS3 A0A0J7L7A3 J3JUI6 A0A2S2R3I0 A0A0C9RMG8 A0A224XJM7 A0A0K8TRD6 A0A182F9F6 A0A1J1IKF5 A0A151ICF6 A0A023F2K1 A0A151XAG3 F4WZV9 A0A158NGN4 A0A2A3EDL4 A0A088ARG9 T1HN22 A0A154P662 J9K1B7 A0A0M9A6H3 E2BAR1 A0A1Y1N351 A0A1Y1N4C0 A0A1B6I177 A0A0L7RIJ5 E0VZL1 A0A1B6CZL8 A0A0A9Z8Z2 A0A1B6GJJ7 A0A1L8D7K1 K7J6K1
Pubmed
19121390
26227816
22651552
22118469
26354079
23622113
+ More
17519023 17510324 24945155 26483478 17994087 17550304 15632085 25315136 20920257 23761445 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 12364791 14747013 17210077 24495485 24845553 25348373 25830018 29403074 25244985 18362917 19820115 24508170 30249741 22516182 23537049 26369729 25474469 21719571 21347285 20798317 28004739 20566863 25401762 26823975 20075255
17519023 17510324 24945155 26483478 17994087 17550304 15632085 25315136 20920257 23761445 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 12364791 14747013 17210077 24495485 24845553 25348373 25830018 29403074 25244985 18362917 19820115 24508170 30249741 22516182 23537049 26369729 25474469 21719571 21347285 20798317 28004739 20566863 25401762 26823975 20075255
EMBL
BABH01002534
ODYU01008491
SOQ52139.1
NWSH01006966
PCG63173.1
JTDY01001580
+ More
KOB73466.1 AK402535 BAM19157.1 AGBW02009983 OWR49406.1 KQ460615 KPJ13591.1 RSAL01000187 RVE44771.1 GAIX01003310 JAA89250.1 KQ459602 KPI93346.1 GFDF01010939 JAV03145.1 EF173369 CH477331 CH477325 ABM68593.1 EAT43322.1 EAT43478.1 JXUM01035858 GAPW01004072 GAPW01004071 GAPW01004070 KQ561076 JAC09528.1 KXJ79790.1 CH940648 EDW61233.1 CM000157 EDW89835.1 CM000071 EAL24642.1 GAPW01004069 JAC09529.1 KA645487 AFP60116.1 AXCM01001575 CH480841 EDW49526.1 ADMH02000438 ETN66491.1 CM002911 KMY91638.1 CH479181 EDW31283.1 JRES01000960 KNC26714.1 AY071708 AAL49330.1 AE013599 BT128775 AAF57321.3 AEK82630.1 AAAB01008844 EAA06065.4 APCN01003108 CH954177 EDV59876.2 GGFM01003514 MBW24265.1 GGFJ01009089 MBW58230.1 GGFK01007142 MBW40463.1 GGFK01006551 MBW39872.1 CH933808 EDW09729.1 AXCN02001148 GAMC01007399 GAMC01007397 JAB99156.1 CP012524 ALC40678.1 KK852591 KDR20697.1 NEVH01021187 PNF20149.1 CH963719 EDW72091.1 DS232130 EDS35655.1 GFDL01007243 JAV27802.1 GDHF01025806 JAI26508.1 JXJN01008672 GANO01003809 JAB56062.1 GAKP01008953 JAC49999.1 GBXI01005770 JAD08522.1 CH916367 EDW01434.1 CH902620 EDV31430.1 PYGN01000538 PSN44594.1 KQ971343 EFA04210.1 KK107063 QOIP01000005 EZA61195.1 RLU22530.1 LBMM01000397 KMQ98460.1 APGK01051818 BT126900 KB741207 KB632327 AEE61862.1 ENN72888.1 ERL92503.1 GGMS01015388 MBY84591.1 GBYB01009525 JAG79292.1 GFTR01003730 JAW12696.1 GDAI01001118 JAI16485.1 CVRI01000048 CRK98937.1 KQ978053 KYM97767.1 GBBI01003172 JAC15540.1 KQ982339 KYQ57354.1 GL888480 EGI60249.1 ADTU01015148 KZ288282 PBC29574.1 ACPB03015126 KQ434826 KZC07426.1 ABLF02010290 KQ435726 KOX78230.1 GL446831 EFN87196.1 GEZM01013932 JAV92259.1 GEZM01013933 JAV92258.1 GECU01027046 JAS80660.1 KQ414584 KOC70623.1 DS235853 EEB18817.1 GEDC01018391 JAS18907.1 GBHO01002703 GBHO01002702 GBHO01002700 GBHO01002699 GDHC01009282 JAG40901.1 JAG40902.1 JAG40904.1 JAG40905.1 JAQ09347.1 GECZ01007306 JAS62463.1 GFDF01011780 JAV02304.1 AAZX01001310
KOB73466.1 AK402535 BAM19157.1 AGBW02009983 OWR49406.1 KQ460615 KPJ13591.1 RSAL01000187 RVE44771.1 GAIX01003310 JAA89250.1 KQ459602 KPI93346.1 GFDF01010939 JAV03145.1 EF173369 CH477331 CH477325 ABM68593.1 EAT43322.1 EAT43478.1 JXUM01035858 GAPW01004072 GAPW01004071 GAPW01004070 KQ561076 JAC09528.1 KXJ79790.1 CH940648 EDW61233.1 CM000157 EDW89835.1 CM000071 EAL24642.1 GAPW01004069 JAC09529.1 KA645487 AFP60116.1 AXCM01001575 CH480841 EDW49526.1 ADMH02000438 ETN66491.1 CM002911 KMY91638.1 CH479181 EDW31283.1 JRES01000960 KNC26714.1 AY071708 AAL49330.1 AE013599 BT128775 AAF57321.3 AEK82630.1 AAAB01008844 EAA06065.4 APCN01003108 CH954177 EDV59876.2 GGFM01003514 MBW24265.1 GGFJ01009089 MBW58230.1 GGFK01007142 MBW40463.1 GGFK01006551 MBW39872.1 CH933808 EDW09729.1 AXCN02001148 GAMC01007399 GAMC01007397 JAB99156.1 CP012524 ALC40678.1 KK852591 KDR20697.1 NEVH01021187 PNF20149.1 CH963719 EDW72091.1 DS232130 EDS35655.1 GFDL01007243 JAV27802.1 GDHF01025806 JAI26508.1 JXJN01008672 GANO01003809 JAB56062.1 GAKP01008953 JAC49999.1 GBXI01005770 JAD08522.1 CH916367 EDW01434.1 CH902620 EDV31430.1 PYGN01000538 PSN44594.1 KQ971343 EFA04210.1 KK107063 QOIP01000005 EZA61195.1 RLU22530.1 LBMM01000397 KMQ98460.1 APGK01051818 BT126900 KB741207 KB632327 AEE61862.1 ENN72888.1 ERL92503.1 GGMS01015388 MBY84591.1 GBYB01009525 JAG79292.1 GFTR01003730 JAW12696.1 GDAI01001118 JAI16485.1 CVRI01000048 CRK98937.1 KQ978053 KYM97767.1 GBBI01003172 JAC15540.1 KQ982339 KYQ57354.1 GL888480 EGI60249.1 ADTU01015148 KZ288282 PBC29574.1 ACPB03015126 KQ434826 KZC07426.1 ABLF02010290 KQ435726 KOX78230.1 GL446831 EFN87196.1 GEZM01013932 JAV92259.1 GEZM01013933 JAV92258.1 GECU01027046 JAS80660.1 KQ414584 KOC70623.1 DS235853 EEB18817.1 GEDC01018391 JAS18907.1 GBHO01002703 GBHO01002702 GBHO01002700 GBHO01002699 GDHC01009282 JAG40901.1 JAG40902.1 JAG40904.1 JAG40905.1 JAQ09347.1 GECZ01007306 JAS62463.1 GFDF01011780 JAV02304.1 AAZX01001310
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053240
UP000283053
+ More
UP000053268 UP000008820 UP000069940 UP000249989 UP000008792 UP000075920 UP000002282 UP000001819 UP000095301 UP000075883 UP000001292 UP000000673 UP000075900 UP000075881 UP000008744 UP000075885 UP000075903 UP000037069 UP000000803 UP000075882 UP000075902 UP000076407 UP000007062 UP000075840 UP000095300 UP000008711 UP000091820 UP000009192 UP000075886 UP000092553 UP000027135 UP000235965 UP000007798 UP000002320 UP000092445 UP000078200 UP000092443 UP000092460 UP000075880 UP000001070 UP000007801 UP000192221 UP000245037 UP000075884 UP000076408 UP000007266 UP000192223 UP000053097 UP000279307 UP000036403 UP000019118 UP000030742 UP000069272 UP000183832 UP000078542 UP000075809 UP000007755 UP000005205 UP000242457 UP000005203 UP000015103 UP000076502 UP000007819 UP000053105 UP000008237 UP000053825 UP000009046 UP000002358
UP000053268 UP000008820 UP000069940 UP000249989 UP000008792 UP000075920 UP000002282 UP000001819 UP000095301 UP000075883 UP000001292 UP000000673 UP000075900 UP000075881 UP000008744 UP000075885 UP000075903 UP000037069 UP000000803 UP000075882 UP000075902 UP000076407 UP000007062 UP000075840 UP000095300 UP000008711 UP000091820 UP000009192 UP000075886 UP000092553 UP000027135 UP000235965 UP000007798 UP000002320 UP000092445 UP000078200 UP000092443 UP000092460 UP000075880 UP000001070 UP000007801 UP000192221 UP000245037 UP000075884 UP000076408 UP000007266 UP000192223 UP000053097 UP000279307 UP000036403 UP000019118 UP000030742 UP000069272 UP000183832 UP000078542 UP000075809 UP000007755 UP000005205 UP000242457 UP000005203 UP000015103 UP000076502 UP000007819 UP000053105 UP000008237 UP000053825 UP000009046 UP000002358
ProteinModelPortal
H9JE01
A0A2H1WGC9
A0A2A4IVM1
A0A0L7LE33
I4DML7
A0A212F6P1
+ More
A0A194RCF9 A0A437B2Q1 S4PMX5 A0A194PIU8 A0A1L8D9V5 Q17AR3 A0A023EK76 B4LPH4 A0A182VXT9 B4NZC2 Q293G9 A0A023EMI3 T1P9U2 A0A182MVR1 B4II00 W5JQV5 A0A182R5Y2 A0A0J9R6A3 A0A182JPE8 B4GBI8 A0A182P773 A0A182VBD2 A0A0L0C5L0 Q8SY89 A1Z6H0 A0A182LAN6 A0A182TTI9 A0A182WV45 Q7PS72 A0A182I8R9 A0A1I8PZ66 B3N3J4 A0A2M3Z6Y5 A0A2M4BYT4 A0A1A9X4R2 A0A2M4AI51 A0A2M4AGN5 B4KLR9 A0A182Q7Z8 W8C0J3 A0A0M4E413 A0A067RA88 A0A2J7PUY9 B4MJ02 B0WVP8 A0A1Q3FJX5 A0A1B0A3P9 A0A1A9V241 A0A0K8UIY8 A0A1A9XQS1 A0A1B0B5C3 U5EFK3 A0A034W5J7 A0A0A1XC51 A0A182J731 B4J9F3 B3MJU1 A0A1W4W7V9 A0A2P8YK55 A0A182NFW1 A0A182XXC8 D6WM26 A0A1W4W956 A0A026WYS3 A0A0J7L7A3 J3JUI6 A0A2S2R3I0 A0A0C9RMG8 A0A224XJM7 A0A0K8TRD6 A0A182F9F6 A0A1J1IKF5 A0A151ICF6 A0A023F2K1 A0A151XAG3 F4WZV9 A0A158NGN4 A0A2A3EDL4 A0A088ARG9 T1HN22 A0A154P662 J9K1B7 A0A0M9A6H3 E2BAR1 A0A1Y1N351 A0A1Y1N4C0 A0A1B6I177 A0A0L7RIJ5 E0VZL1 A0A1B6CZL8 A0A0A9Z8Z2 A0A1B6GJJ7 A0A1L8D7K1 K7J6K1
A0A194RCF9 A0A437B2Q1 S4PMX5 A0A194PIU8 A0A1L8D9V5 Q17AR3 A0A023EK76 B4LPH4 A0A182VXT9 B4NZC2 Q293G9 A0A023EMI3 T1P9U2 A0A182MVR1 B4II00 W5JQV5 A0A182R5Y2 A0A0J9R6A3 A0A182JPE8 B4GBI8 A0A182P773 A0A182VBD2 A0A0L0C5L0 Q8SY89 A1Z6H0 A0A182LAN6 A0A182TTI9 A0A182WV45 Q7PS72 A0A182I8R9 A0A1I8PZ66 B3N3J4 A0A2M3Z6Y5 A0A2M4BYT4 A0A1A9X4R2 A0A2M4AI51 A0A2M4AGN5 B4KLR9 A0A182Q7Z8 W8C0J3 A0A0M4E413 A0A067RA88 A0A2J7PUY9 B4MJ02 B0WVP8 A0A1Q3FJX5 A0A1B0A3P9 A0A1A9V241 A0A0K8UIY8 A0A1A9XQS1 A0A1B0B5C3 U5EFK3 A0A034W5J7 A0A0A1XC51 A0A182J731 B4J9F3 B3MJU1 A0A1W4W7V9 A0A2P8YK55 A0A182NFW1 A0A182XXC8 D6WM26 A0A1W4W956 A0A026WYS3 A0A0J7L7A3 J3JUI6 A0A2S2R3I0 A0A0C9RMG8 A0A224XJM7 A0A0K8TRD6 A0A182F9F6 A0A1J1IKF5 A0A151ICF6 A0A023F2K1 A0A151XAG3 F4WZV9 A0A158NGN4 A0A2A3EDL4 A0A088ARG9 T1HN22 A0A154P662 J9K1B7 A0A0M9A6H3 E2BAR1 A0A1Y1N351 A0A1Y1N4C0 A0A1B6I177 A0A0L7RIJ5 E0VZL1 A0A1B6CZL8 A0A0A9Z8Z2 A0A1B6GJJ7 A0A1L8D7K1 K7J6K1
Ontologies
GO
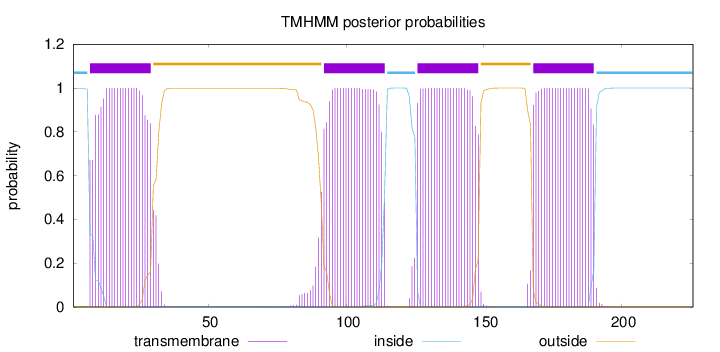
Topology
Length:
226
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
91.8971
Exp number, first 60 AAs:
22.63528
Total prob of N-in:
0.99892
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 91
TMhelix
92 - 114
inside
115 - 125
TMhelix
126 - 148
outside
149 - 167
TMhelix
168 - 190
inside
191 - 226
Population Genetic Test Statistics
Pi
155.907758
Theta
153.461964
Tajima's D
0.050106
CLR
0
CSRT
0.385430728463577
Interpretation
Uncertain
