Gene
KWMTBOMO08991 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007783
Annotation
PREDICTED:_succinyl-CoA_ligase_[ADP-forming]_subunit_beta?_mitochondrial_[Papilio_xuthus]
Full name
Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial
+ More
Succinate-CoA ligase subunit beta
Succinate-CoA ligase subunit beta
Alternative Name
ATP-specific succinyl-CoA synthetase subunit beta
Succinyl-CoA synthetase beta-A chain
Succinyl-CoA synthetase beta-A chain
Location in the cell
Mitochondrial Reliability : 2.165
Sequence
CDS
ATGGCGATTGTATTACCTCGTTCTCTTGGTTTTGCTGAAACAATATTATTTAGATATGGAAATAAGATATTGACAGCATCTTCAGCCAACAAATTTCCAAGTAAACAACAAGTCCGCCACTTGAATGTACATGAATACATAAGTTATACTCTACTACGGGACCATGGAATTCCTGTACCCAAATTTAATGTGGCCAAAACCAAGGATGAGGCAATTAAGTTTGCCACAGAACTCAATACCAAAGACATCGTTCTAAAAGCACAGGTTCTTGCTGGTGGTCGAGGGAAAGGTACCTTTAAGAATGGACTTAAAGGTGGAGTTCGAATGGTCAACACACCTGAAGTTGCTGGTGATATAGCAGGAAAGATGCTTAAACAACTCCTAGTAACAAAACAAACAGGGGCAGCAGGACGAATTTGCAACATGGTCATGGTCACAGAGAGGAAGTTCCCACGCAGGGAATACTACGTGGCAATTATGATGGAACGCAGTTTCAATGGTCCAGTCATCATTGCTTCATCTCAAGGTGGTGTCAACATTGAAGATGTTGCAGCTGAAAATCCTGATGCAATAACTTATGAACCTATTGATATTGTTTCGGGTATTACTGATGATCAAATTTGCCGTGTTATCGAAAAGATCGGTCTACAAGAATTTGCAACTGAAGCTCATGGTATGATTAAAAAAATGTATGATTTGTTCCTTAAGAAGGATGCCTTATTGATTGAAGTAAATCCGTATGCCGAAGATGCCTTGACAGGACAATTCTTCTGTTTGGATGCCAAGTTTAGATTTGACGATAATGCTGAATTCAGACAAAAAGAGCTATTTAGTTTGAGAGACACTACTCAAGAAGACCCCAAGGAAATTGAAGCAGCTAAATACAACTTAAATTACATTGCTCTCGATGGAAACATTGGGTGTATGGTTAACGGTGCTGGATTAGCTATGGCTACGATGGATATAATCAAACTCTACGGGGGTGACCCCGCTAACTTCTTGGATGTCGGTGGAGGCGCTACTGCCCAGGCTGTCTCTGAGGCGTTCAAAATCATTCTCTCAGACCCGAAGGTGACAGCTATTTTGGTGAACATTTTTGGTGGTATTATGAGGTGCGACGTCATTGCAGAAGGTATCATTAATGCTGCCAAAAATCTGAATATACAGATCCCAGTGATTGTTCGCCTTCAAGGAACGAAGGTTAACGAAGCTAGAAAGTTGGTAGCCGAGTCCGGGCTTCGCATCGTGCCTCGCGACGACCTGGACGAGGCCGCCAAACTCGCCGTGCAACTCTCAGAGATCGTTGCCCTAGCCAGAAAGGCTGGAGTTGAGGTTAAATTTGATATTCCAAGAGTTATGCTTGAAAAGTAA
Protein
MAIVLPRSLGFAETILFRYGNKILTASSANKFPSKQQVRHLNVHEYISYTLLRDHGIPVPKFNVAKTKDEAIKFATELNTKDIVLKAQVLAGGRGKGTFKNGLKGGVRMVNTPEVAGDIAGKMLKQLLVTKQTGAAGRICNMVMVTERKFPRREYYVAIMMERSFNGPVIIASSQGGVNIEDVAAENPDAITYEPIDIVSGITDDQICRVIEKIGLQEFATEAHGMIKKMYDLFLKKDALLIEVNPYAEDALTGQFFCLDAKFRFDDNAEFRQKELFSLRDTTQEDPKEIEAAKYNLNYIALDGNIGCMVNGAGLAMATMDIIKLYGGDPANFLDVGGGATAQAVSEAFKIILSDPKVTAILVNIFGGIMRCDVIAEGIINAAKNLNIQIPVIVRLQGTKVNEARKLVAESGLRIVPRDDLDEAAKLAVQLSEIVALARKAGVEVKFDIPRVMLEK
Summary
Description
ATP-specific succinyl-CoA synthetase functions in the citric acid cycle (TCA), coupling the hydrolysis of succinyl-CoA to the synthesis of ATP and thus represents the only step of substrate-level phosphorylation in the TCA. The beta subunit provides nucleotide specificity of the enzyme and binds the substrate succinate, while the binding sites for coenzyme A and phosphate are found in the alpha subunit.
Catalytic Activity
ATP + CoA + succinate = ADP + phosphate + succinyl-CoA
Cofactor
Mg(2+)
Subunit
Heterodimer of an alpha and a beta subunit. The beta subunit determines specificity for ATP.
Heterodimer of an alpha and a beta subunit.
Heterodimer of an alpha and a beta subunit.
Similarity
Belongs to the succinate/malate CoA ligase beta subunit family. ATP-specific subunit beta subfamily.
Belongs to the succinate/malate CoA ligase beta subunit family.
Belongs to the succinate/malate CoA ligase beta subunit family.
Feature
chain Succinate-CoA ligase subunit beta
Uniprot
A0A194PKT6
A0A194R7V5
S4NSU9
A0A212F6Q3
A0A2H1WGI7
A0A2A4IVF8
+ More
H9JE36 W8BJB4 W8BZS1 A0A034VMK8 A0A2J7QIN4 A0A067R278 A0A0L0CLN1 A0A1I8MEN7 A0A2J7QVC5 D6WNP7 A0A0P4VNG2 A0A0T6AU44 T1PF72 A0A1L8EGZ6 A0A023FCI6 A0A1L8EGY7 A0A1B6FWG5 J3JT86 Q16P82 A0A0V0G4C3 D3TM34 A0A069DUI1 A0A1B0C0H3 A0A1A9YDY8 A0A1A9Z6E2 A0A182HMZ8 A0A023EUP7 A0A1A9VA18 A0A1B6HGI8 A0A182QTV2 K7IUH8 A0A0A9X9V3 A0A182VNQ9 A0A182LI78 Q7PMT2 A0A224XNB4 A0A232F6W6 T1HFZ7 A0A1B6E4X5 A0A084VZT2 Q16P83 A0A182MX75 A0A182JEX9 A0A182NNB2 A0A182R2R2 A0A023ESD2 A0A0R3NJ42 A0A182P605 A0A182WAR1 A0A0R3NPJ5 A0A1W4W933 A0A182WZW1 A0A1A9WH09 B0WFW7 A0A0M5IZT8 N6T7J6 A0A1L8E0M1 B5DYI6 A0A336MYF9 T1DE80 A0A2M3ZFY6 A0A0Q9WFA8 A0A0Q9X7F6 A0A1L8E0U5 A0A3B0KDT3 A0A1Q3FK18 B4GFU8 A0A1Q3FK72 A0A182YI33 A0A0R1E1S6 A0A0Q9WNB3 B4LXK7 A0A126GUR6 A0A1W4W320 B4N817 A0A0C9RA06 Q95U38 B4K9E6 A0A0Q9X0L0 A0A0P8XXK4 B4HKZ0 A0A182FZQ3 A0A0N8P1A9 A0A1J1IZ15 R4WE04 B4JTJ3 A0A2M4AP58 A0A2M4BN71 B4PTX7 A0A195C7K0 B3P1W3 A0A0B4JCW4 A0A2M4A401 A0A1W4VQZ0
H9JE36 W8BJB4 W8BZS1 A0A034VMK8 A0A2J7QIN4 A0A067R278 A0A0L0CLN1 A0A1I8MEN7 A0A2J7QVC5 D6WNP7 A0A0P4VNG2 A0A0T6AU44 T1PF72 A0A1L8EGZ6 A0A023FCI6 A0A1L8EGY7 A0A1B6FWG5 J3JT86 Q16P82 A0A0V0G4C3 D3TM34 A0A069DUI1 A0A1B0C0H3 A0A1A9YDY8 A0A1A9Z6E2 A0A182HMZ8 A0A023EUP7 A0A1A9VA18 A0A1B6HGI8 A0A182QTV2 K7IUH8 A0A0A9X9V3 A0A182VNQ9 A0A182LI78 Q7PMT2 A0A224XNB4 A0A232F6W6 T1HFZ7 A0A1B6E4X5 A0A084VZT2 Q16P83 A0A182MX75 A0A182JEX9 A0A182NNB2 A0A182R2R2 A0A023ESD2 A0A0R3NJ42 A0A182P605 A0A182WAR1 A0A0R3NPJ5 A0A1W4W933 A0A182WZW1 A0A1A9WH09 B0WFW7 A0A0M5IZT8 N6T7J6 A0A1L8E0M1 B5DYI6 A0A336MYF9 T1DE80 A0A2M3ZFY6 A0A0Q9WFA8 A0A0Q9X7F6 A0A1L8E0U5 A0A3B0KDT3 A0A1Q3FK18 B4GFU8 A0A1Q3FK72 A0A182YI33 A0A0R1E1S6 A0A0Q9WNB3 B4LXK7 A0A126GUR6 A0A1W4W320 B4N817 A0A0C9RA06 Q95U38 B4K9E6 A0A0Q9X0L0 A0A0P8XXK4 B4HKZ0 A0A182FZQ3 A0A0N8P1A9 A0A1J1IZ15 R4WE04 B4JTJ3 A0A2M4AP58 A0A2M4BN71 B4PTX7 A0A195C7K0 B3P1W3 A0A0B4JCW4 A0A2M4A401 A0A1W4VQZ0
EC Number
6.2.1.5
6.2.1.-
6.2.1.-
Pubmed
26354079
23622113
22118469
19121390
24495485
25348373
+ More
24845553 26108605 25315136 18362917 19820115 27129103 25474469 22516182 23537049 17510324 20353571 26334808 24945155 20075255 25401762 26823975 20966253 12364791 14747013 17210077 28648823 24438588 15632085 17994087 24330624 18057021 25244985 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23691247
24845553 26108605 25315136 18362917 19820115 27129103 25474469 22516182 23537049 17510324 20353571 26334808 24945155 20075255 25401762 26823975 20966253 12364791 14747013 17210077 28648823 24438588 15632085 17994087 24330624 18057021 25244985 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23691247
EMBL
KQ459602
KPI93344.1
KQ460615
KPJ13589.1
GAIX01013902
JAA78658.1
+ More
AGBW02009983 OWR49404.1 ODYU01008491 SOQ52137.1 NWSH01006966 PCG63172.1 BABH01002535 BABH01002536 GAMC01007723 JAB98832.1 GAMC01007724 JAB98831.1 GAKP01014381 GAKP01014379 JAC44571.1 NEVH01013583 PNF28442.1 KK852751 KDR17135.1 JRES01000223 KNC33205.1 NEVH01010477 PNF32534.1 KQ971342 EFA03750.1 GDKW01000414 JAI56181.1 LJIG01022813 KRT78584.1 KA647442 AFP62071.1 GFDG01000812 JAV17987.1 GBBI01000214 JAC18498.1 GFDG01000822 JAV17977.1 GECZ01015223 JAS54546.1 BT126442 KB630727 KB632357 AEE61406.1 ERL83818.1 ERL93444.1 CH477792 EAT36152.1 GECL01003227 JAP02897.1 CCAG010009033 EZ422486 ADD18762.1 GBGD01001428 JAC87461.1 JXJN01023622 APCN01008727 GAPW01001414 JAC12184.1 GECU01033862 GECU01013174 JAS73844.1 JAS94532.1 AXCN02001904 AAZX01002659 GBHO01035289 GBHO01029724 GBRD01008202 GBRD01004408 GDHC01010966 JAG08315.1 JAG13880.1 JAG57619.1 JAQ07663.1 AAAB01008968 EAA13239.4 GFTR01006354 JAW10072.1 NNAY01000861 OXU26169.1 ACPB03014676 GEDC01004344 JAS32954.1 ATLV01018955 KE525256 KFB43476.1 EAT36151.1 AXCM01000075 GAPW01001435 JAC12163.1 CM000070 KRT00916.1 KRT00915.1 DS231921 EDS26502.1 CP012526 ALC46361.1 APGK01040619 KB740984 ENN76189.1 GFDF01001833 JAV12251.1 EDY68366.2 UFQS01002857 UFQT01002857 SSX14777.1 SSX34169.1 GALA01001147 JAA93705.1 GGFM01006599 MBW27350.1 CH940650 KRF82930.1 CH933806 KRG01626.1 GFDF01001800 JAV12284.1 OUUW01000008 SPP84429.1 GFDL01007167 JAV27878.1 CH479182 EDW34483.1 GFDL01007152 JAV27893.1 CM000160 KRK03216.1 KRF82929.1 EDW66791.1 AE014297 ALI30546.1 CH964232 EDW81268.2 GBYB01003651 JAG73418.1 AY058331 AAL13560.1 ACZ94855.1 EDW15578.2 KRG01627.1 CH902617 KPU79409.1 CH480815 EDW42954.1 KPU79412.1 CVRI01000064 CRL05523.1 AK418009 BAN21224.1 CH916373 EDV95083.1 GGFK01009232 MBW42553.1 GGFJ01005321 MBW54462.1 EDW96588.1 KQ978231 KYM96128.1 CH954181 EDV49851.1 ADV37289.2 GGFK01002131 MBW35452.1
AGBW02009983 OWR49404.1 ODYU01008491 SOQ52137.1 NWSH01006966 PCG63172.1 BABH01002535 BABH01002536 GAMC01007723 JAB98832.1 GAMC01007724 JAB98831.1 GAKP01014381 GAKP01014379 JAC44571.1 NEVH01013583 PNF28442.1 KK852751 KDR17135.1 JRES01000223 KNC33205.1 NEVH01010477 PNF32534.1 KQ971342 EFA03750.1 GDKW01000414 JAI56181.1 LJIG01022813 KRT78584.1 KA647442 AFP62071.1 GFDG01000812 JAV17987.1 GBBI01000214 JAC18498.1 GFDG01000822 JAV17977.1 GECZ01015223 JAS54546.1 BT126442 KB630727 KB632357 AEE61406.1 ERL83818.1 ERL93444.1 CH477792 EAT36152.1 GECL01003227 JAP02897.1 CCAG010009033 EZ422486 ADD18762.1 GBGD01001428 JAC87461.1 JXJN01023622 APCN01008727 GAPW01001414 JAC12184.1 GECU01033862 GECU01013174 JAS73844.1 JAS94532.1 AXCN02001904 AAZX01002659 GBHO01035289 GBHO01029724 GBRD01008202 GBRD01004408 GDHC01010966 JAG08315.1 JAG13880.1 JAG57619.1 JAQ07663.1 AAAB01008968 EAA13239.4 GFTR01006354 JAW10072.1 NNAY01000861 OXU26169.1 ACPB03014676 GEDC01004344 JAS32954.1 ATLV01018955 KE525256 KFB43476.1 EAT36151.1 AXCM01000075 GAPW01001435 JAC12163.1 CM000070 KRT00916.1 KRT00915.1 DS231921 EDS26502.1 CP012526 ALC46361.1 APGK01040619 KB740984 ENN76189.1 GFDF01001833 JAV12251.1 EDY68366.2 UFQS01002857 UFQT01002857 SSX14777.1 SSX34169.1 GALA01001147 JAA93705.1 GGFM01006599 MBW27350.1 CH940650 KRF82930.1 CH933806 KRG01626.1 GFDF01001800 JAV12284.1 OUUW01000008 SPP84429.1 GFDL01007167 JAV27878.1 CH479182 EDW34483.1 GFDL01007152 JAV27893.1 CM000160 KRK03216.1 KRF82929.1 EDW66791.1 AE014297 ALI30546.1 CH964232 EDW81268.2 GBYB01003651 JAG73418.1 AY058331 AAL13560.1 ACZ94855.1 EDW15578.2 KRG01627.1 CH902617 KPU79409.1 CH480815 EDW42954.1 KPU79412.1 CVRI01000064 CRL05523.1 AK418009 BAN21224.1 CH916373 EDV95083.1 GGFK01009232 MBW42553.1 GGFJ01005321 MBW54462.1 EDW96588.1 KQ978231 KYM96128.1 CH954181 EDV49851.1 ADV37289.2 GGFK01002131 MBW35452.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000218220
UP000005204
UP000235965
+ More
UP000027135 UP000037069 UP000095301 UP000007266 UP000030742 UP000008820 UP000092444 UP000092460 UP000092443 UP000092445 UP000075840 UP000078200 UP000075886 UP000002358 UP000075903 UP000075882 UP000007062 UP000215335 UP000015103 UP000030765 UP000075883 UP000075880 UP000075884 UP000075900 UP000001819 UP000075885 UP000075920 UP000192223 UP000076407 UP000091820 UP000002320 UP000092553 UP000019118 UP000008792 UP000009192 UP000268350 UP000008744 UP000076408 UP000002282 UP000000803 UP000192221 UP000007798 UP000007801 UP000001292 UP000069272 UP000183832 UP000001070 UP000078542 UP000008711
UP000027135 UP000037069 UP000095301 UP000007266 UP000030742 UP000008820 UP000092444 UP000092460 UP000092443 UP000092445 UP000075840 UP000078200 UP000075886 UP000002358 UP000075903 UP000075882 UP000007062 UP000215335 UP000015103 UP000030765 UP000075883 UP000075880 UP000075884 UP000075900 UP000001819 UP000075885 UP000075920 UP000192223 UP000076407 UP000091820 UP000002320 UP000092553 UP000019118 UP000008792 UP000009192 UP000268350 UP000008744 UP000076408 UP000002282 UP000000803 UP000192221 UP000007798 UP000007801 UP000001292 UP000069272 UP000183832 UP000001070 UP000078542 UP000008711
PRIDE
Interpro
SUPFAM
SSF52210
SSF52210
Gene 3D
ProteinModelPortal
A0A194PKT6
A0A194R7V5
S4NSU9
A0A212F6Q3
A0A2H1WGI7
A0A2A4IVF8
+ More
H9JE36 W8BJB4 W8BZS1 A0A034VMK8 A0A2J7QIN4 A0A067R278 A0A0L0CLN1 A0A1I8MEN7 A0A2J7QVC5 D6WNP7 A0A0P4VNG2 A0A0T6AU44 T1PF72 A0A1L8EGZ6 A0A023FCI6 A0A1L8EGY7 A0A1B6FWG5 J3JT86 Q16P82 A0A0V0G4C3 D3TM34 A0A069DUI1 A0A1B0C0H3 A0A1A9YDY8 A0A1A9Z6E2 A0A182HMZ8 A0A023EUP7 A0A1A9VA18 A0A1B6HGI8 A0A182QTV2 K7IUH8 A0A0A9X9V3 A0A182VNQ9 A0A182LI78 Q7PMT2 A0A224XNB4 A0A232F6W6 T1HFZ7 A0A1B6E4X5 A0A084VZT2 Q16P83 A0A182MX75 A0A182JEX9 A0A182NNB2 A0A182R2R2 A0A023ESD2 A0A0R3NJ42 A0A182P605 A0A182WAR1 A0A0R3NPJ5 A0A1W4W933 A0A182WZW1 A0A1A9WH09 B0WFW7 A0A0M5IZT8 N6T7J6 A0A1L8E0M1 B5DYI6 A0A336MYF9 T1DE80 A0A2M3ZFY6 A0A0Q9WFA8 A0A0Q9X7F6 A0A1L8E0U5 A0A3B0KDT3 A0A1Q3FK18 B4GFU8 A0A1Q3FK72 A0A182YI33 A0A0R1E1S6 A0A0Q9WNB3 B4LXK7 A0A126GUR6 A0A1W4W320 B4N817 A0A0C9RA06 Q95U38 B4K9E6 A0A0Q9X0L0 A0A0P8XXK4 B4HKZ0 A0A182FZQ3 A0A0N8P1A9 A0A1J1IZ15 R4WE04 B4JTJ3 A0A2M4AP58 A0A2M4BN71 B4PTX7 A0A195C7K0 B3P1W3 A0A0B4JCW4 A0A2M4A401 A0A1W4VQZ0
H9JE36 W8BJB4 W8BZS1 A0A034VMK8 A0A2J7QIN4 A0A067R278 A0A0L0CLN1 A0A1I8MEN7 A0A2J7QVC5 D6WNP7 A0A0P4VNG2 A0A0T6AU44 T1PF72 A0A1L8EGZ6 A0A023FCI6 A0A1L8EGY7 A0A1B6FWG5 J3JT86 Q16P82 A0A0V0G4C3 D3TM34 A0A069DUI1 A0A1B0C0H3 A0A1A9YDY8 A0A1A9Z6E2 A0A182HMZ8 A0A023EUP7 A0A1A9VA18 A0A1B6HGI8 A0A182QTV2 K7IUH8 A0A0A9X9V3 A0A182VNQ9 A0A182LI78 Q7PMT2 A0A224XNB4 A0A232F6W6 T1HFZ7 A0A1B6E4X5 A0A084VZT2 Q16P83 A0A182MX75 A0A182JEX9 A0A182NNB2 A0A182R2R2 A0A023ESD2 A0A0R3NJ42 A0A182P605 A0A182WAR1 A0A0R3NPJ5 A0A1W4W933 A0A182WZW1 A0A1A9WH09 B0WFW7 A0A0M5IZT8 N6T7J6 A0A1L8E0M1 B5DYI6 A0A336MYF9 T1DE80 A0A2M3ZFY6 A0A0Q9WFA8 A0A0Q9X7F6 A0A1L8E0U5 A0A3B0KDT3 A0A1Q3FK18 B4GFU8 A0A1Q3FK72 A0A182YI33 A0A0R1E1S6 A0A0Q9WNB3 B4LXK7 A0A126GUR6 A0A1W4W320 B4N817 A0A0C9RA06 Q95U38 B4K9E6 A0A0Q9X0L0 A0A0P8XXK4 B4HKZ0 A0A182FZQ3 A0A0N8P1A9 A0A1J1IZ15 R4WE04 B4JTJ3 A0A2M4AP58 A0A2M4BN71 B4PTX7 A0A195C7K0 B3P1W3 A0A0B4JCW4 A0A2M4A401 A0A1W4VQZ0
PDB
6G4Q
E-value=1.06121e-142,
Score=1299
Ontologies
PATHWAY
GO
PANTHER
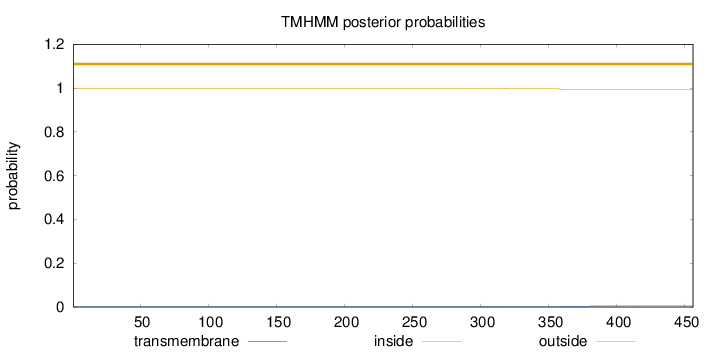
Topology
Subcellular location
Mitochondrion
Length:
456
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14738
Exp number, first 60 AAs:
0.00113
Total prob of N-in:
0.00196
outside
1 - 456
Population Genetic Test Statistics
Pi
207.373671
Theta
166.124734
Tajima's D
0.763454
CLR
0.338741
CSRT
0.587020648967552
Interpretation
Uncertain
