Gene
KWMTBOMO08988
Pre Gene Modal
BGIBMGA007785
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_multidrug_resistance-associated_protein_4_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.705
Sequence
CDS
ATGGAGTCAAAAGTAGTGTTGAACAATAAGAATCCTCACGATGAAGCTAATATCCTGTCGAAGACGTTTTTAACGTGGAGTTTTACTTTATTCAAAAGGGGTTATAAACAGGGCATTTCTATCGACGACTTATGGCAGGCTCGTGGGGCAGATCATTCGAAGCAACTCGGCGATCGTCTTGAAGTAGCCTGGGAAAGAGAGTTAGAAAGAGCCAAACAGACTGGTACAAAACCATCTTTCTCGAAAGCGATCATTCGCTCATTCTGGCTCGAATATATGATTTGCGGCATATTTGTTGGTTTTCTCTTTATCGTATTATGGCCACTGGTTCCGTTTACACTAGCATTGTTTATCGGATATTTCTCTGGAGAAAAATCACCTGAAAATTATAAAAACGCTCACATCTACAATTTCTTGATGAACTTTTTGAGTATAACGACGTCTTTAATGTTAAATCATTTACAACTTAGTCAGGGGAGAGTCGGTATGCGGATAAGGATTGCTAGCTGCTCGTTACTATATAGAAAGATTCTCAAACTTGACAGGACAGGATTAGCTAAAACAGAGCCCGGTCAAGTGATTAATCTGATGTCAAACGATGTTAATCGATTTGATTTGGTTGTTTTGTTTTTGAATTACATCTGGGTAATGCCTATTGTTGTACCTGTTGTTTCGTATCTCGTCTGGCAGCATATAGGCTGGGCCACACTTGCAGCGCTCATTGTAATATTTTTACAAACAGTTGTCGTGCAAGCGTATCTAAGTAATATGCAAGGTGTTCTTCGTGGAAAAATTGCGAAACGCACCGATGAGAGAGTAAAAGTTATGAGCGAATTGGTGAACGGAGTTCAAGTAATCAAAATGTATGCTTGGGAAAAACCGTTTGAGAAACTCGTTGATAAACTGAGAAAGCTCGAAGTGCATTACATAATGCGAACGTCCATGATCAAAGGTTTTTCAACAGCACTGAGCGTGTTCACGGAGCGTTTCATACTTTTCGCGGCCATCGTCGCATTCGTAGTGATGGGGGGCGAAATACGTTCAGAAATAACTTTCTCATTAGTGCAGTACTTCAATCTTTTGCAACTCGCCTGTAACATATTCTTTCCCTTGGCTTTAGCATTTCTTGCTGAGTCAAAAGTCTCAGTGAGTCGTTTGGAGGAATTTCTGTTGTTGGACGAGTTAGAACAAACTAAGCAAAATGAGCCTATCAAAGCCGCAATGGACTCTAAACTCATTTTGACCAATGGAAACGCAAAAGACACTGAGAAAATTAAATCGAAATCAACTGGTTTAAGTTTGACCGGAGTTTCGGCCAGTTGGTCAACTGATCCAATAGTCCACACATTGAGAAACATTACGTTTTCCGCTGAACCAGGAGAATTTGTTGGGATCGCCGGTCTCGTTGGTGCCGGTAAGTCGTCTCTGCTACAAGTGCTCCTGGGAGAACTGAAGCCATCTCAGGGGACCGTATCATTGGATGGTGCGCGTATTTCTTACGCAAGTCAAGAGCCGTGGCTCTTCGTGGCGACGATTAGACAGAACATCTTGTTCGGCTTACCATACGATAAAACACGTTACAAGAAGGTGGTGACTGCGTGCGCTCTGCTGCGGGACTTCGAGCAGTTGCCGGCGGGAGACTGCACGCTGGTCGGCGAGCGCGGCACCAGTCTCAGCGGGGGGCAGCGGGCGCGGGTGGGACTCGCGCGTGCCTGCTACAGACAAGCCGACATATATCTCCTCGACGATCCATTGTCAGCGGTGGACACGCATGTTGGCAAACATCTCGTTTCCGAATGTGTGAATGGATTGCTGCAGAACCACACCCGCATTCTGGTCACACATCAGTTGCATCACCTCAAAACCGCTGATAAAGTCATCATTCTGCGTAATGGAGAAATTGAAATGCAAGGAAAATTCGAAGAGGCGTCACGCTGTCCACTCTTTTCTGAAATGGAAGAGGAAGATGAACCCGATGAAAAAAATCTAAAACATCTCAGGAAACGAACTATCTCCATTCAGTCCAAAACGAGTGTTAGTACAACACACGATGGAAGCCAGGTTGATGTTGAAGAAGATGAGCAGAAGGAGTCCGAAGAACTAACTGGATCTGGGAGAGTATCAGGATTAGTATACATGAGCTACTTCCGTGCCGGCGGTGGTTGGGCACTTTTGTTCTGGACTTTGTTTTCCATCATTATTGCACAAGCCGTGACGTCAATGAGCGATATGTGGCTCACACACTGGATGAACGACGTGGAAGTGACGTACTTGACACCGACTTTTGAAACGGATCTCCTTACTGATCGCAACGAAACGAATAATTTAAATTTAACGAGTGAAGTTCCCACAGTCGTTGACTCGGCCGCCTTCCCGCCAAACTTAACAGTGATCGGCACGATCATGAAAACGGCCATGGCCATACAGAACGCGACGGAAACGGAACGTGAAAATGTAAATCACTCGTATTACATTTACATATGGGCGATCGGAATATTGGGTTGCATAATATTAACAACGGCCAGATCGATGATGTTCCTGTGGGTGTGTATGCGGAGCTCTATAAAACTCCACAACCAAATGTTCAGTAATATTATCGCGGCGACGATGCGCTTCTTCGACACAAACCCGTCCGGACGAGTTCTCAACAGATTCTCGAAGGACATGGGCGTAGTCGACGAGATACTGCCCCGGATGTTTTTAGACAGCATACAGATATTTATGGTGATGATCGGTATATTAGTGATGGTCGCTATAGTGAGCCCCTTCATGATGCTTACGACGGTAGTGTGCGGGGTGCTCATGTATCTGTGGACCGTCGTTTACCTCACCACCGCTCAAGCTGTAAAAAGGGTCGAAGGCATATCTCGGAGCCCTGTGTTCTCCCACGTGTCGGCCAGCATGGCGGGCCTGTCCACCGTGAGGGCTTGTAACGCTGAAATGATGCTGGCGGATCAATTCGACGATAAGCAAGACGTACACACAGCCGCCTGGTATTTAACTCTGGTCACAAACACTGCTTTCTCGATCTGGCTCAGTCTCATATCTGCCTTGTACGTTATAGTTGTGGCCTATACATTTTTACTACTAGATGACGGAACAACGAAATCTGGAAACGTCGGTCTGGCGTTAAGTCAAGGATTAATTTTGGTGAACATGGTTCAGTATGGTATCAAACAGACAACGGAGGTTATATCGCAAATGACCAGCGTCGAAAGAGTGATACAATTCACGTCGCTGCCTCTCGAGAAGACGGAAGGGCCCGCCCCGCCGCCGAGCTGGCCGCAGCGCGCTCGCGTCGTATTCAAGGATCTCAATCTTCGATACGACAAAGATTCCGACCCGGTGCTCAGAAACCTTAACATCATCATTGAAAGCGGCTGGAAGGTCGGTGTCGTCGGGCGCACCGGTGCCGGTAAATCATCATTGATCTCGGCTTTGTTCCGATTGGCTCCGATCGAGGGTCATATTTATATCGATGATGTTGAAACTGGAGAAATCGCTCTTAAAGCATTGCGCTCGAAGATTTCCATCATTCCTCAAGAACCAGTCCTGTTTTCTGCTACTTTGCGATACAATTTGGATCCATTCGATAAATACACTGACGCTGAAATATGGACCGCCCTAGAACAGGTTGAACTCAAAAATACTGTGACGTCATTATCATCATCAGTCGCTTCTGGTGGATCCAATTTTAGTGCTGGACAACGGCAGCTTCTGTGCCTGGCCCGAGCGGCCCTTGCTAGAAATAGACTACTCGTGTTGGATGAAGCTACCGCTAATGTAGATCCGAATACTGATGCACTGATCCAGAAGTCTATTCGCAAACATTTTGCGGACTGCACCGTTGTCACAGTCGCCCATCGACTACACACCGTTGCGGATTCTGATCGAGTTGTGGTCATGGAAGCAGGCCAGATCGTCGAATGTGGCCACCCTTACGAGCTACTTAAGAACGATAACGGGCATTTCTCGAAAATGGTTAAACAATTGGGTCCCGCCTCAGAGCAGAGCCTTAGAGAACTTGCGCGGGACGCACACGCACAACACATACAATACGTCGACGCGGACGACCAGGATAAAACGAAATAA
Protein
MESKVVLNNKNPHDEANILSKTFLTWSFTLFKRGYKQGISIDDLWQARGADHSKQLGDRLEVAWERELERAKQTGTKPSFSKAIIRSFWLEYMICGIFVGFLFIVLWPLVPFTLALFIGYFSGEKSPENYKNAHIYNFLMNFLSITTSLMLNHLQLSQGRVGMRIRIASCSLLYRKILKLDRTGLAKTEPGQVINLMSNDVNRFDLVVLFLNYIWVMPIVVPVVSYLVWQHIGWATLAALIVIFLQTVVVQAYLSNMQGVLRGKIAKRTDERVKVMSELVNGVQVIKMYAWEKPFEKLVDKLRKLEVHYIMRTSMIKGFSTALSVFTERFILFAAIVAFVVMGGEIRSEITFSLVQYFNLLQLACNIFFPLALAFLAESKVSVSRLEEFLLLDELEQTKQNEPIKAAMDSKLILTNGNAKDTEKIKSKSTGLSLTGVSASWSTDPIVHTLRNITFSAEPGEFVGIAGLVGAGKSSLLQVLLGELKPSQGTVSLDGARISYASQEPWLFVATIRQNILFGLPYDKTRYKKVVTACALLRDFEQLPAGDCTLVGERGTSLSGGQRARVGLARACYRQADIYLLDDPLSAVDTHVGKHLVSECVNGLLQNHTRILVTHQLHHLKTADKVIILRNGEIEMQGKFEEASRCPLFSEMEEEDEPDEKNLKHLRKRTISIQSKTSVSTTHDGSQVDVEEDEQKESEELTGSGRVSGLVYMSYFRAGGGWALLFWTLFSIIIAQAVTSMSDMWLTHWMNDVEVTYLTPTFETDLLTDRNETNNLNLTSEVPTVVDSAAFPPNLTVIGTIMKTAMAIQNATETERENVNHSYYIYIWAIGILGCIILTTARSMMFLWVCMRSSIKLHNQMFSNIIAATMRFFDTNPSGRVLNRFSKDMGVVDEILPRMFLDSIQIFMVMIGILVMVAIVSPFMMLTTVVCGVLMYLWTVVYLTTAQAVKRVEGISRSPVFSHVSASMAGLSTVRACNAEMMLADQFDDKQDVHTAAWYLTLVTNTAFSIWLSLISALYVIVVAYTFLLLDDGTTKSGNVGLALSQGLILVNMVQYGIKQTTEVISQMTSVERVIQFTSLPLEKTEGPAPPPSWPQRARVVFKDLNLRYDKDSDPVLRNLNIIIESGWKVGVVGRTGAGKSSLISALFRLAPIEGHIYIDDVETGEIALKALRSKISIIPQEPVLFSATLRYNLDPFDKYTDAEIWTALEQVELKNTVTSLSSSVASGGSNFSAGQRQLLCLARAALARNRLLVLDEATANVDPNTDALIQKSIRKHFADCTVVTVAHRLHTVADSDRVVVMEAGQIVECGHPYELLKNDNGHFSKMVKQLGPASEQSLRELARDAHAQHIQYVDADDQDKTK
Summary
Uniprot
A0A0E3ZB25
A0A194PIU3
A0A2W1BFZ5
D2K6M7
A0A2A4K224
D2K6M6
+ More
A0A437B2W7 A0A2A4J643 A0A0E3UVD8 A0A194RCF4 A0A2A4K3R8 A0A1Y1KY01 D6WLS5 A0A0U9HSH2 A0A0C9R3H3 A0A067R1W1 A0A1W4WE54 A0A0M8ZRW3 A0A154PH22 A0A2J7PFD7 A0A026WLA8 A0A088AFE6 A0A0E3UUF7 A0A3G1PY98 A0A3S2TFR5 V5TF62 A0A151IYU4 D6WJN5 A0A2A3ED65 V5I8K8 U5JE76 V5TEA6 A0A0C9R1Z8 A0A158NMX0 A0A151WHK6 A0A0C9RET5 F4WTF6 A0A195BC45 A0A2H1X0D3 U4URU5 A0A0T6AV71 A0A3L8DIZ7 A0A232EXT2 K7IUA5 A0A2J7PFE3 A0A1W4WP56 A0A232F6R5 A0A195EWQ3 A0A1W4WE28 A0A146LYI9 A0A2J7PEN6 A0A0A9WJ51 A0A023F3H7 A0A0L7QUH9 A0A158NLT2 A0A2J7PVQ7 A0A336K0I7 A0A067R9U7 A0A1Y1LFL8 A0A0P4W3Q5 A0A182MAA3 K7IUA6 A0A182NJ65 A0A1W5PZ91 Q178E8 A0A182WC63 A0A067RQX2 A0A0V0G5R1 A0A182GUU2 A0A336LRH9 A0A182Q5F7 A0A1Y1M6K8 A0A069DYA9 A0A1Y1M4A8 J9KIT1 A0A1A9V725 A0A182RML1 A0A0L0CKR5 A0A182VRI3 A0A2J7R104 A0A1L8DV41 Q7Q5I0 A0A1L8DUP6 A0A1B0FJ40 W8AYC4 A0A1S4GS60 A0A182MS57 A0A182V3W3 A0A182TI84 A0A182I405 A0A182VF60 A0A2J7QE18 A0A182LCH3 A0A1I8JUN0 A0A067RUE2 A0A182HJA2
A0A437B2W7 A0A2A4J643 A0A0E3UVD8 A0A194RCF4 A0A2A4K3R8 A0A1Y1KY01 D6WLS5 A0A0U9HSH2 A0A0C9R3H3 A0A067R1W1 A0A1W4WE54 A0A0M8ZRW3 A0A154PH22 A0A2J7PFD7 A0A026WLA8 A0A088AFE6 A0A0E3UUF7 A0A3G1PY98 A0A3S2TFR5 V5TF62 A0A151IYU4 D6WJN5 A0A2A3ED65 V5I8K8 U5JE76 V5TEA6 A0A0C9R1Z8 A0A158NMX0 A0A151WHK6 A0A0C9RET5 F4WTF6 A0A195BC45 A0A2H1X0D3 U4URU5 A0A0T6AV71 A0A3L8DIZ7 A0A232EXT2 K7IUA5 A0A2J7PFE3 A0A1W4WP56 A0A232F6R5 A0A195EWQ3 A0A1W4WE28 A0A146LYI9 A0A2J7PEN6 A0A0A9WJ51 A0A023F3H7 A0A0L7QUH9 A0A158NLT2 A0A2J7PVQ7 A0A336K0I7 A0A067R9U7 A0A1Y1LFL8 A0A0P4W3Q5 A0A182MAA3 K7IUA6 A0A182NJ65 A0A1W5PZ91 Q178E8 A0A182WC63 A0A067RQX2 A0A0V0G5R1 A0A182GUU2 A0A336LRH9 A0A182Q5F7 A0A1Y1M6K8 A0A069DYA9 A0A1Y1M4A8 J9KIT1 A0A1A9V725 A0A182RML1 A0A0L0CKR5 A0A182VRI3 A0A2J7R104 A0A1L8DV41 Q7Q5I0 A0A1L8DUP6 A0A1B0FJ40 W8AYC4 A0A1S4GS60 A0A182MS57 A0A182V3W3 A0A182TI84 A0A182I405 A0A182VF60 A0A2J7QE18 A0A182LCH3 A0A1I8JUN0 A0A067RUE2 A0A182HJA2
Pubmed
EMBL
KM245563
AKC96182.1
KQ459602
KPI93341.1
KZ150089
PZC73708.1
+ More
GU182416 ACZ64280.1 NWSH01000215 PCG78321.1 GU182415 ACZ64279.1 RSAL01000187 RVE44780.1 NWSH01003134 PCG66893.1 KM245564 AKC96183.1 KQ460615 KPJ13586.1 PCG78320.1 GEZM01073523 JAV65030.1 KQ971343 EFA04157.2 GARF01000038 JAC88908.1 GBYB01007337 JAG77104.1 KK853083 KDR11623.1 KQ435879 KOX69880.1 KQ434902 KZC11097.1 NEVH01025663 PNF15046.1 KK107159 EZA56728.1 KM245560 AKC96179.1 KY701524 AVV62497.1 RVE44791.1 KF278997 AHB63235.1 KQ980745 KYN13667.1 EFA04472.2 KZ288287 PBC29424.1 GALX01004510 JAB63956.1 KC112554 AGN29368.1 KF278996 AHB63234.1 GBYB01006857 JAG76624.1 ADTU01020829 KQ983115 KYQ47297.1 GBYB01006860 JAG76627.1 GL888334 EGI62548.1 KQ976529 KYM81765.1 ODYU01012016 SOQ58094.1 KB632335 ERL92805.1 LJIG01022728 KRT79020.1 QOIP01000007 RLU20387.1 NNAY01001666 OXU23276.1 AAZX01003864 PNF15053.1 NNAY01000861 OXU26170.1 KQ981940 KYN32705.1 GDHC01006604 JAQ12025.1 NEVH01026090 PNF14801.1 GBHO01036168 JAG07436.1 GBBI01002866 JAC15846.1 KQ414735 KOC62298.1 ADTU01019804 NEVH01020943 PNF20422.1 UFQS01000040 UFQT01000040 SSW98269.1 SSX18655.1 KK852607 KDR20353.1 GEZM01061568 JAV70366.1 GDKW01000545 JAI56050.1 AXCM01002561 KY250427 APG42670.1 CH477365 EAT42553.1 KK852521 KDR22129.1 GECL01002731 JAP03393.1 JXUM01089709 JXUM01089710 UFQS01000118 UFQT01000118 SSX00015.1 SSX20395.1 AXCN02000455 GEZM01041017 JAV80648.1 GBGD01000127 JAC88762.1 GEZM01041016 JAV80649.1 ABLF02035624 JRES01000264 KNC32832.1 NEVH01008214 PNF34507.1 GFDF01003929 JAV10155.1 AAAB01008960 EAA10720.3 GFDF01003930 JAV10154.1 CCAG010022756 GAMC01020689 GAMC01020688 GAMC01020687 JAB85866.1 AXCM01014272 APCN01002262 APCN01002263 NEVH01015354 PNF26826.1 KK852413 KDR24430.1 APCN01002134 APCN01002135
GU182416 ACZ64280.1 NWSH01000215 PCG78321.1 GU182415 ACZ64279.1 RSAL01000187 RVE44780.1 NWSH01003134 PCG66893.1 KM245564 AKC96183.1 KQ460615 KPJ13586.1 PCG78320.1 GEZM01073523 JAV65030.1 KQ971343 EFA04157.2 GARF01000038 JAC88908.1 GBYB01007337 JAG77104.1 KK853083 KDR11623.1 KQ435879 KOX69880.1 KQ434902 KZC11097.1 NEVH01025663 PNF15046.1 KK107159 EZA56728.1 KM245560 AKC96179.1 KY701524 AVV62497.1 RVE44791.1 KF278997 AHB63235.1 KQ980745 KYN13667.1 EFA04472.2 KZ288287 PBC29424.1 GALX01004510 JAB63956.1 KC112554 AGN29368.1 KF278996 AHB63234.1 GBYB01006857 JAG76624.1 ADTU01020829 KQ983115 KYQ47297.1 GBYB01006860 JAG76627.1 GL888334 EGI62548.1 KQ976529 KYM81765.1 ODYU01012016 SOQ58094.1 KB632335 ERL92805.1 LJIG01022728 KRT79020.1 QOIP01000007 RLU20387.1 NNAY01001666 OXU23276.1 AAZX01003864 PNF15053.1 NNAY01000861 OXU26170.1 KQ981940 KYN32705.1 GDHC01006604 JAQ12025.1 NEVH01026090 PNF14801.1 GBHO01036168 JAG07436.1 GBBI01002866 JAC15846.1 KQ414735 KOC62298.1 ADTU01019804 NEVH01020943 PNF20422.1 UFQS01000040 UFQT01000040 SSW98269.1 SSX18655.1 KK852607 KDR20353.1 GEZM01061568 JAV70366.1 GDKW01000545 JAI56050.1 AXCM01002561 KY250427 APG42670.1 CH477365 EAT42553.1 KK852521 KDR22129.1 GECL01002731 JAP03393.1 JXUM01089709 JXUM01089710 UFQS01000118 UFQT01000118 SSX00015.1 SSX20395.1 AXCN02000455 GEZM01041017 JAV80648.1 GBGD01000127 JAC88762.1 GEZM01041016 JAV80649.1 ABLF02035624 JRES01000264 KNC32832.1 NEVH01008214 PNF34507.1 GFDF01003929 JAV10155.1 AAAB01008960 EAA10720.3 GFDF01003930 JAV10154.1 CCAG010022756 GAMC01020689 GAMC01020688 GAMC01020687 JAB85866.1 AXCM01014272 APCN01002262 APCN01002263 NEVH01015354 PNF26826.1 KK852413 KDR24430.1 APCN01002134 APCN01002135
Proteomes
UP000053268
UP000218220
UP000283053
UP000053240
UP000007266
UP000027135
+ More
UP000192223 UP000053105 UP000076502 UP000235965 UP000053097 UP000005203 UP000078492 UP000242457 UP000005205 UP000075809 UP000007755 UP000078540 UP000030742 UP000279307 UP000215335 UP000002358 UP000078541 UP000053825 UP000075883 UP000075884 UP000008820 UP000075920 UP000069940 UP000075886 UP000007819 UP000078200 UP000075900 UP000037069 UP000007062 UP000092444 UP000075903 UP000075902 UP000075840 UP000075882
UP000192223 UP000053105 UP000076502 UP000235965 UP000053097 UP000005203 UP000078492 UP000242457 UP000005205 UP000075809 UP000007755 UP000078540 UP000030742 UP000279307 UP000215335 UP000002358 UP000078541 UP000053825 UP000075883 UP000075884 UP000008820 UP000075920 UP000069940 UP000075886 UP000007819 UP000078200 UP000075900 UP000037069 UP000007062 UP000092444 UP000075903 UP000075902 UP000075840 UP000075882
Interpro
Gene 3D
ProteinModelPortal
A0A0E3ZB25
A0A194PIU3
A0A2W1BFZ5
D2K6M7
A0A2A4K224
D2K6M6
+ More
A0A437B2W7 A0A2A4J643 A0A0E3UVD8 A0A194RCF4 A0A2A4K3R8 A0A1Y1KY01 D6WLS5 A0A0U9HSH2 A0A0C9R3H3 A0A067R1W1 A0A1W4WE54 A0A0M8ZRW3 A0A154PH22 A0A2J7PFD7 A0A026WLA8 A0A088AFE6 A0A0E3UUF7 A0A3G1PY98 A0A3S2TFR5 V5TF62 A0A151IYU4 D6WJN5 A0A2A3ED65 V5I8K8 U5JE76 V5TEA6 A0A0C9R1Z8 A0A158NMX0 A0A151WHK6 A0A0C9RET5 F4WTF6 A0A195BC45 A0A2H1X0D3 U4URU5 A0A0T6AV71 A0A3L8DIZ7 A0A232EXT2 K7IUA5 A0A2J7PFE3 A0A1W4WP56 A0A232F6R5 A0A195EWQ3 A0A1W4WE28 A0A146LYI9 A0A2J7PEN6 A0A0A9WJ51 A0A023F3H7 A0A0L7QUH9 A0A158NLT2 A0A2J7PVQ7 A0A336K0I7 A0A067R9U7 A0A1Y1LFL8 A0A0P4W3Q5 A0A182MAA3 K7IUA6 A0A182NJ65 A0A1W5PZ91 Q178E8 A0A182WC63 A0A067RQX2 A0A0V0G5R1 A0A182GUU2 A0A336LRH9 A0A182Q5F7 A0A1Y1M6K8 A0A069DYA9 A0A1Y1M4A8 J9KIT1 A0A1A9V725 A0A182RML1 A0A0L0CKR5 A0A182VRI3 A0A2J7R104 A0A1L8DV41 Q7Q5I0 A0A1L8DUP6 A0A1B0FJ40 W8AYC4 A0A1S4GS60 A0A182MS57 A0A182V3W3 A0A182TI84 A0A182I405 A0A182VF60 A0A2J7QE18 A0A182LCH3 A0A1I8JUN0 A0A067RUE2 A0A182HJA2
A0A437B2W7 A0A2A4J643 A0A0E3UVD8 A0A194RCF4 A0A2A4K3R8 A0A1Y1KY01 D6WLS5 A0A0U9HSH2 A0A0C9R3H3 A0A067R1W1 A0A1W4WE54 A0A0M8ZRW3 A0A154PH22 A0A2J7PFD7 A0A026WLA8 A0A088AFE6 A0A0E3UUF7 A0A3G1PY98 A0A3S2TFR5 V5TF62 A0A151IYU4 D6WJN5 A0A2A3ED65 V5I8K8 U5JE76 V5TEA6 A0A0C9R1Z8 A0A158NMX0 A0A151WHK6 A0A0C9RET5 F4WTF6 A0A195BC45 A0A2H1X0D3 U4URU5 A0A0T6AV71 A0A3L8DIZ7 A0A232EXT2 K7IUA5 A0A2J7PFE3 A0A1W4WP56 A0A232F6R5 A0A195EWQ3 A0A1W4WE28 A0A146LYI9 A0A2J7PEN6 A0A0A9WJ51 A0A023F3H7 A0A0L7QUH9 A0A158NLT2 A0A2J7PVQ7 A0A336K0I7 A0A067R9U7 A0A1Y1LFL8 A0A0P4W3Q5 A0A182MAA3 K7IUA6 A0A182NJ65 A0A1W5PZ91 Q178E8 A0A182WC63 A0A067RQX2 A0A0V0G5R1 A0A182GUU2 A0A336LRH9 A0A182Q5F7 A0A1Y1M6K8 A0A069DYA9 A0A1Y1M4A8 J9KIT1 A0A1A9V725 A0A182RML1 A0A0L0CKR5 A0A182VRI3 A0A2J7R104 A0A1L8DV41 Q7Q5I0 A0A1L8DUP6 A0A1B0FJ40 W8AYC4 A0A1S4GS60 A0A182MS57 A0A182V3W3 A0A182TI84 A0A182I405 A0A182VF60 A0A2J7QE18 A0A182LCH3 A0A1I8JUN0 A0A067RUE2 A0A182HJA2
PDB
6C3P
E-value=1.91042e-125,
Score=1155
Ontologies
GO
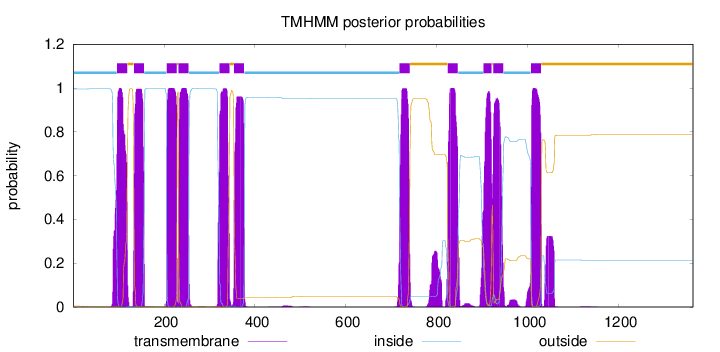
Topology
Length:
1363
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
254.6684
Exp number, first 60 AAs:
0.01775
Total prob of N-in:
0.99524
inside
1 - 96
TMhelix
97 - 119
outside
120 - 133
TMhelix
134 - 156
inside
157 - 205
TMhelix
206 - 228
outside
229 - 231
TMhelix
232 - 254
inside
255 - 321
TMhelix
322 - 344
outside
345 - 353
TMhelix
354 - 376
inside
377 - 717
TMhelix
718 - 740
outside
741 - 823
TMhelix
824 - 846
inside
847 - 902
TMhelix
903 - 920
outside
921 - 923
TMhelix
924 - 946
inside
947 - 1006
TMhelix
1007 - 1029
outside
1030 - 1363
Population Genetic Test Statistics
Pi
222.950311
Theta
176.53918
Tajima's D
0.761123
CLR
0.747847
CSRT
0.587120643967802
Interpretation
Uncertain
