Gene
KWMTBOMO08987
Pre Gene Modal
BGIBMGA007744
Annotation
PREDICTED:_DNA_replication_complex_GINS_protein_SLD5_[Amyelois_transitella]
Full name
DNA replication complex GINS protein SLD5
Location in the cell
Nuclear Reliability : 2.92
Sequence
CDS
ATGGAGTCAACCGAAGAGTTGCAGTTCAGTGATGATGAAGAAGAAGTAACAGCTGAGACTGTTTTAAAAACACTACAAAGTGCTTGGCAAAATGAAAGATTGTCCCCAGAAATACTCCCTCATCAAAATGACATGGTAGAATGTATGTTAGGTCAAATACAACACATGGAAAGAAACATAAATAAACTTTCAAAAACTGATTTGAGAACTTCTATACACAAAATGGAGATTAGCAGAATTAAATTTATTATTTGTAATTATCTTAAAACTAGACTGAATAAGATTGAAAAATATTGCATTCCAATACTAAACCAAGAAAGACAAAGAATAGAAACTGATACTAATTACCTAACACCATCAGAATATAAATATGCTCAAGAATATGTTTTGAACATGGAAAATCATATGAAAAATGTGGTATTAAATAAAATGCCTGGTAATATGCAAGTCTTTGAAATGAACAAAATGGCAGAACATCCTAATTTACAAACGCATGTATTCCTTAAAGCCAATGAAACCATTAATGGGGTTGTATTAGAAGACTTGACTGGTGAAGATGAAGAAATTGACTTAGAAGAGGGCTCGCAGCACATCTTGCAATACAAACCAATTGCAGATCTTTTAAAGAACGGGAAAGTCACATTGTTATGA
Protein
MESTEELQFSDDEEEVTAETVLKTLQSAWQNERLSPEILPHQNDMVECMLGQIQHMERNINKLSKTDLRTSIHKMEISRIKFIICNYLKTRLNKIEKYCIPILNQERQRIETDTNYLTPSEYKYAQEYVLNMENHMKNVVLNKMPGNMQVFEMNKMAEHPNLQTHVFLKANETINGVVLEDLTGEDEEIDLEEGSQHILQYKPIADLLKNGKVTLL
Summary
Description
The GINS complex plays an essential role in the initiation of DNA replication.
Similarity
Belongs to the GINS4/SLD5 family.
Uniprot
A0A2H1X1Y0
A0A2A4J5T9
A0A1E1W923
S4PE03
A0A3S2LUP1
A0A194PQD0
+ More
A0A212FAL0 A0A194R8L0 A0A0L7LDW1 A0A158NMW9 A0A067QQL3 A0A195BB37 A0A195EXU9 A0A151IYP1 F4WTF5 A0A195C5J0 A0A026WKZ4 A0A2J7PFE2 A0A3L8E2R1 A0A0J7KB58 A0A151WHL0 A0A0N0BD34 E2AHJ0 E9IWB6 E2C456 A0A2A3ECB9 A0A154PIF3 A0A0L7QKT8 A0A2H8TLL6 A0A1B6I1Q1 K7J574 A0A232ERX6 C4WT18 A0A1B6EQ23 A0A1B6E5G4 A0A2P8YA37 A0A433T9L9 A7SEX8 V9LAY9 A0A3B3RWW1 A0A1S3I1A7 A0A224YND5 A0A3R7Q6B5 W5NGR2 W5UBN9 W5KD60 A0A2Y9FMQ6 A0A3P8YKL2 A0A210PHN6 A0A1W4Z723 A0A3M6V6F8 A0A1S3D044 A0A287BEE8 A0A3B4ED28 A0A384A473 A0A452J6B7 A0A3Q2YIR5 A0A131Z4K9 A0A131XLK3 A0A131Y546 A0A2R8Q9M1 A0A3Q1JKL8 A0A0P7TSJ4 A0A341D1Q8 A0A3B4X869 A0A3B4VDZ8 A0A2Y9MYN2 A0A2U4A8P8 Q6DC44 G3PAY9 A0A060WY14 A0A340X6B7 A0A402EJB1 H9G413 F6XB21 G3PAY7 A0A3B4GI03 A0A3Q2VI21 A0A3Q4HMS7 A0A3P8PD73 A0A3P9BYY3 A0A1E1XFM3 A0A2B4SK78 A0A3B5BDP0 A0A1A8G281 A0A1A8I1R0 A0A1A8C6X8 A0A1A8AL57 A0A3Q1HGD0 A0A3Q1ALI8 A0A3P8T4R5 A0A1B0GJX3 A0A3B4BBB3 I3JWY1 A0A023FG22 A0A091VCW5 A0A2U9BRI2 A0A1A8QHU8 A0A3Q3LSG9 A0A2D4NKZ3
A0A212FAL0 A0A194R8L0 A0A0L7LDW1 A0A158NMW9 A0A067QQL3 A0A195BB37 A0A195EXU9 A0A151IYP1 F4WTF5 A0A195C5J0 A0A026WKZ4 A0A2J7PFE2 A0A3L8E2R1 A0A0J7KB58 A0A151WHL0 A0A0N0BD34 E2AHJ0 E9IWB6 E2C456 A0A2A3ECB9 A0A154PIF3 A0A0L7QKT8 A0A2H8TLL6 A0A1B6I1Q1 K7J574 A0A232ERX6 C4WT18 A0A1B6EQ23 A0A1B6E5G4 A0A2P8YA37 A0A433T9L9 A7SEX8 V9LAY9 A0A3B3RWW1 A0A1S3I1A7 A0A224YND5 A0A3R7Q6B5 W5NGR2 W5UBN9 W5KD60 A0A2Y9FMQ6 A0A3P8YKL2 A0A210PHN6 A0A1W4Z723 A0A3M6V6F8 A0A1S3D044 A0A287BEE8 A0A3B4ED28 A0A384A473 A0A452J6B7 A0A3Q2YIR5 A0A131Z4K9 A0A131XLK3 A0A131Y546 A0A2R8Q9M1 A0A3Q1JKL8 A0A0P7TSJ4 A0A341D1Q8 A0A3B4X869 A0A3B4VDZ8 A0A2Y9MYN2 A0A2U4A8P8 Q6DC44 G3PAY9 A0A060WY14 A0A340X6B7 A0A402EJB1 H9G413 F6XB21 G3PAY7 A0A3B4GI03 A0A3Q2VI21 A0A3Q4HMS7 A0A3P8PD73 A0A3P9BYY3 A0A1E1XFM3 A0A2B4SK78 A0A3B5BDP0 A0A1A8G281 A0A1A8I1R0 A0A1A8C6X8 A0A1A8AL57 A0A3Q1HGD0 A0A3Q1ALI8 A0A3P8T4R5 A0A1B0GJX3 A0A3B4BBB3 I3JWY1 A0A023FG22 A0A091VCW5 A0A2U9BRI2 A0A1A8QHU8 A0A3Q3LSG9 A0A2D4NKZ3
Pubmed
23622113
26354079
22118469
26227816
21347285
24845553
+ More
21719571 24508170 30249741 20798317 21282665 20075255 28648823 29403074 17615350 24402279 29240929 28797301 23127152 25329095 25069045 28812685 30382153 30723633 28562605 26830274 28049606 23594743 24755649 21881562 18464734 25186727 28503490 25463417
21719571 24508170 30249741 20798317 21282665 20075255 28648823 29403074 17615350 24402279 29240929 28797301 23127152 25329095 25069045 28812685 30382153 30723633 28562605 26830274 28049606 23594743 24755649 21881562 18464734 25186727 28503490 25463417
EMBL
ODYU01012831
SOQ59331.1
NWSH01003134
PCG66894.1
GDQN01007693
JAT83361.1
+ More
GAIX01003411 JAA89149.1 RSAL01000187 RVE44779.1 KQ459602 KPI93340.1 AGBW02009451 OWR50776.1 KQ460615 KPJ13585.1 JTDY01001605 KOB73391.1 ADTU01020829 KK853083 KDR11621.1 KQ976529 KYM81766.1 KQ981940 KYN32704.1 KQ980745 KYN13669.1 GL888334 EGI62547.1 KQ978231 KYM96119.1 KK107159 EZA56727.1 NEVH01025663 PNF15054.1 QOIP01000001 RLU26994.1 LBMM01010369 KMQ87509.1 KQ983115 KYQ47298.1 KQ435879 KOX69881.1 GL439548 EFN67093.1 GL766486 EFZ15131.1 GL452396 EFN77285.1 KZ288287 PBC29423.1 KQ434902 KZC11098.1 KQ414940 KOC59209.1 GFXV01000937 GFXV01003124 MBW12742.1 MBW14929.1 GECU01026882 JAS80824.1 AAZX01000336 NNAY01002545 OXU21091.1 ABLF02027798 AK340425 BAH71038.1 GECZ01029740 JAS40029.1 GEDC01031352 GEDC01004122 GEDC01003420 JAS05946.1 JAS33176.1 JAS33878.1 PYGN01000761 PSN41122.1 RQTK01000523 RUS78246.1 DS469640 EDO37714.1 JW876810 AFP09327.1 GFPF01007972 MAA19118.1 QCYY01002528 ROT69702.1 AHAT01008509 JT411270 AHH39363.1 NEDP02076689 OWF35991.1 RCHS01000010 RMX61461.1 AEMK02000105 DQIR01095044 DQIR01208250 DQIR01215759 DQIR01218694 DQIR01220794 DQIR01251833 DQIR01263257 HDA50520.1 GEDV01002243 JAP86314.1 GEFH01000197 JAP68384.1 GEFM01002630 JAP73166.1 CABZ01070468 JARO02010440 KPP60690.1 BC078242 BC131877 AAH78242.1 FR904670 CDQ69490.1 BDOT01000015 GCF44305.1 AAWZ02030553 AAPN01429195 AAPN01429196 GFAC01001146 JAT98042.1 LSMT01000060 PFX29776.1 HAEB01017947 SBQ64474.1 HAED01004736 SBQ90766.1 HADZ01011403 HAEA01008077 SBP75344.1 HADY01017284 HAEJ01007092 SBP55769.1 AJWK01022430 AERX01013610 GBBK01003551 JAC20931.1 KL410942 KFR01212.1 CP026251 AWP06818.1 HAEG01012854 SBR93415.1 IACN01006882 IACN01006883 IACN01006884 IACN01006885 IACN01006886 IACN01006887 IACN01006888 LAB46391.1
GAIX01003411 JAA89149.1 RSAL01000187 RVE44779.1 KQ459602 KPI93340.1 AGBW02009451 OWR50776.1 KQ460615 KPJ13585.1 JTDY01001605 KOB73391.1 ADTU01020829 KK853083 KDR11621.1 KQ976529 KYM81766.1 KQ981940 KYN32704.1 KQ980745 KYN13669.1 GL888334 EGI62547.1 KQ978231 KYM96119.1 KK107159 EZA56727.1 NEVH01025663 PNF15054.1 QOIP01000001 RLU26994.1 LBMM01010369 KMQ87509.1 KQ983115 KYQ47298.1 KQ435879 KOX69881.1 GL439548 EFN67093.1 GL766486 EFZ15131.1 GL452396 EFN77285.1 KZ288287 PBC29423.1 KQ434902 KZC11098.1 KQ414940 KOC59209.1 GFXV01000937 GFXV01003124 MBW12742.1 MBW14929.1 GECU01026882 JAS80824.1 AAZX01000336 NNAY01002545 OXU21091.1 ABLF02027798 AK340425 BAH71038.1 GECZ01029740 JAS40029.1 GEDC01031352 GEDC01004122 GEDC01003420 JAS05946.1 JAS33176.1 JAS33878.1 PYGN01000761 PSN41122.1 RQTK01000523 RUS78246.1 DS469640 EDO37714.1 JW876810 AFP09327.1 GFPF01007972 MAA19118.1 QCYY01002528 ROT69702.1 AHAT01008509 JT411270 AHH39363.1 NEDP02076689 OWF35991.1 RCHS01000010 RMX61461.1 AEMK02000105 DQIR01095044 DQIR01208250 DQIR01215759 DQIR01218694 DQIR01220794 DQIR01251833 DQIR01263257 HDA50520.1 GEDV01002243 JAP86314.1 GEFH01000197 JAP68384.1 GEFM01002630 JAP73166.1 CABZ01070468 JARO02010440 KPP60690.1 BC078242 BC131877 AAH78242.1 FR904670 CDQ69490.1 BDOT01000015 GCF44305.1 AAWZ02030553 AAPN01429195 AAPN01429196 GFAC01001146 JAT98042.1 LSMT01000060 PFX29776.1 HAEB01017947 SBQ64474.1 HAED01004736 SBQ90766.1 HADZ01011403 HAEA01008077 SBP75344.1 HADY01017284 HAEJ01007092 SBP55769.1 AJWK01022430 AERX01013610 GBBK01003551 JAC20931.1 KL410942 KFR01212.1 CP026251 AWP06818.1 HAEG01012854 SBR93415.1 IACN01006882 IACN01006883 IACN01006884 IACN01006885 IACN01006886 IACN01006887 IACN01006888 LAB46391.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000005205 UP000027135 UP000078540 UP000078541 UP000078492 UP000007755 UP000078542 UP000053097 UP000235965 UP000279307 UP000036403 UP000075809 UP000053105 UP000000311 UP000008237 UP000242457 UP000076502 UP000053825 UP000002358 UP000215335 UP000007819 UP000245037 UP000271974 UP000001593 UP000261540 UP000085678 UP000283509 UP000018468 UP000221080 UP000018467 UP000248484 UP000265140 UP000242188 UP000192224 UP000275408 UP000079169 UP000008227 UP000261440 UP000261681 UP000291020 UP000264820 UP000000437 UP000265040 UP000034805 UP000252040 UP000261360 UP000261420 UP000248483 UP000245320 UP000007635 UP000193380 UP000265300 UP000288954 UP000001646 UP000002279 UP000261460 UP000264840 UP000261580 UP000265100 UP000265160 UP000225706 UP000261400 UP000257200 UP000257160 UP000265080 UP000092461 UP000261520 UP000005207 UP000053283 UP000246464 UP000261640
UP000005205 UP000027135 UP000078540 UP000078541 UP000078492 UP000007755 UP000078542 UP000053097 UP000235965 UP000279307 UP000036403 UP000075809 UP000053105 UP000000311 UP000008237 UP000242457 UP000076502 UP000053825 UP000002358 UP000215335 UP000007819 UP000245037 UP000271974 UP000001593 UP000261540 UP000085678 UP000283509 UP000018468 UP000221080 UP000018467 UP000248484 UP000265140 UP000242188 UP000192224 UP000275408 UP000079169 UP000008227 UP000261440 UP000261681 UP000291020 UP000264820 UP000000437 UP000265040 UP000034805 UP000252040 UP000261360 UP000261420 UP000248483 UP000245320 UP000007635 UP000193380 UP000265300 UP000288954 UP000001646 UP000002279 UP000261460 UP000264840 UP000261580 UP000265100 UP000265160 UP000225706 UP000261400 UP000257200 UP000257160 UP000265080 UP000092461 UP000261520 UP000005207 UP000053283 UP000246464 UP000261640
Interpro
SUPFAM
SSF158573
SSF158573
ProteinModelPortal
A0A2H1X1Y0
A0A2A4J5T9
A0A1E1W923
S4PE03
A0A3S2LUP1
A0A194PQD0
+ More
A0A212FAL0 A0A194R8L0 A0A0L7LDW1 A0A158NMW9 A0A067QQL3 A0A195BB37 A0A195EXU9 A0A151IYP1 F4WTF5 A0A195C5J0 A0A026WKZ4 A0A2J7PFE2 A0A3L8E2R1 A0A0J7KB58 A0A151WHL0 A0A0N0BD34 E2AHJ0 E9IWB6 E2C456 A0A2A3ECB9 A0A154PIF3 A0A0L7QKT8 A0A2H8TLL6 A0A1B6I1Q1 K7J574 A0A232ERX6 C4WT18 A0A1B6EQ23 A0A1B6E5G4 A0A2P8YA37 A0A433T9L9 A7SEX8 V9LAY9 A0A3B3RWW1 A0A1S3I1A7 A0A224YND5 A0A3R7Q6B5 W5NGR2 W5UBN9 W5KD60 A0A2Y9FMQ6 A0A3P8YKL2 A0A210PHN6 A0A1W4Z723 A0A3M6V6F8 A0A1S3D044 A0A287BEE8 A0A3B4ED28 A0A384A473 A0A452J6B7 A0A3Q2YIR5 A0A131Z4K9 A0A131XLK3 A0A131Y546 A0A2R8Q9M1 A0A3Q1JKL8 A0A0P7TSJ4 A0A341D1Q8 A0A3B4X869 A0A3B4VDZ8 A0A2Y9MYN2 A0A2U4A8P8 Q6DC44 G3PAY9 A0A060WY14 A0A340X6B7 A0A402EJB1 H9G413 F6XB21 G3PAY7 A0A3B4GI03 A0A3Q2VI21 A0A3Q4HMS7 A0A3P8PD73 A0A3P9BYY3 A0A1E1XFM3 A0A2B4SK78 A0A3B5BDP0 A0A1A8G281 A0A1A8I1R0 A0A1A8C6X8 A0A1A8AL57 A0A3Q1HGD0 A0A3Q1ALI8 A0A3P8T4R5 A0A1B0GJX3 A0A3B4BBB3 I3JWY1 A0A023FG22 A0A091VCW5 A0A2U9BRI2 A0A1A8QHU8 A0A3Q3LSG9 A0A2D4NKZ3
A0A212FAL0 A0A194R8L0 A0A0L7LDW1 A0A158NMW9 A0A067QQL3 A0A195BB37 A0A195EXU9 A0A151IYP1 F4WTF5 A0A195C5J0 A0A026WKZ4 A0A2J7PFE2 A0A3L8E2R1 A0A0J7KB58 A0A151WHL0 A0A0N0BD34 E2AHJ0 E9IWB6 E2C456 A0A2A3ECB9 A0A154PIF3 A0A0L7QKT8 A0A2H8TLL6 A0A1B6I1Q1 K7J574 A0A232ERX6 C4WT18 A0A1B6EQ23 A0A1B6E5G4 A0A2P8YA37 A0A433T9L9 A7SEX8 V9LAY9 A0A3B3RWW1 A0A1S3I1A7 A0A224YND5 A0A3R7Q6B5 W5NGR2 W5UBN9 W5KD60 A0A2Y9FMQ6 A0A3P8YKL2 A0A210PHN6 A0A1W4Z723 A0A3M6V6F8 A0A1S3D044 A0A287BEE8 A0A3B4ED28 A0A384A473 A0A452J6B7 A0A3Q2YIR5 A0A131Z4K9 A0A131XLK3 A0A131Y546 A0A2R8Q9M1 A0A3Q1JKL8 A0A0P7TSJ4 A0A341D1Q8 A0A3B4X869 A0A3B4VDZ8 A0A2Y9MYN2 A0A2U4A8P8 Q6DC44 G3PAY9 A0A060WY14 A0A340X6B7 A0A402EJB1 H9G413 F6XB21 G3PAY7 A0A3B4GI03 A0A3Q2VI21 A0A3Q4HMS7 A0A3P8PD73 A0A3P9BYY3 A0A1E1XFM3 A0A2B4SK78 A0A3B5BDP0 A0A1A8G281 A0A1A8I1R0 A0A1A8C6X8 A0A1A8AL57 A0A3Q1HGD0 A0A3Q1ALI8 A0A3P8T4R5 A0A1B0GJX3 A0A3B4BBB3 I3JWY1 A0A023FG22 A0A091VCW5 A0A2U9BRI2 A0A1A8QHU8 A0A3Q3LSG9 A0A2D4NKZ3
PDB
2Q9Q
E-value=1.00818e-37,
Score=390
Ontologies
GO
PANTHER
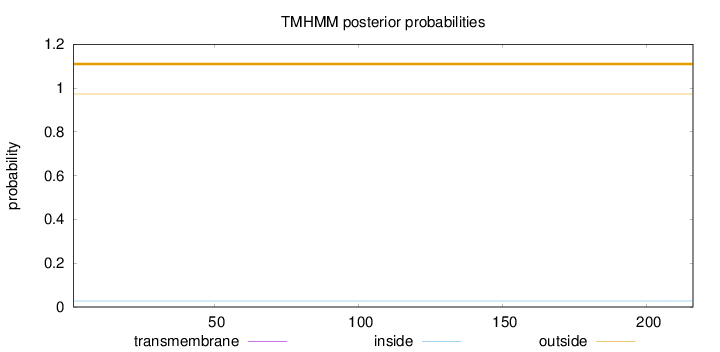
Topology
Subcellular location
Nucleus
Length:
216
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02742
outside
1 - 216
Population Genetic Test Statistics
Pi
299.490478
Theta
196.470316
Tajima's D
1.627965
CLR
0
CSRT
0.813109344532773
Interpretation
Uncertain
