Gene
KWMTBOMO08981
Pre Gene Modal
BGIBMGA007789
Annotation
cryptochrome_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.21
Sequence
CDS
ATGTCAGCAGCCCCAGAAACACTGCCACCACCAAGTGCTCAAGCTCACACGCCGGCCAGGCCTACTCATATGTCCGCACCTCGACGTACGCCGGGTAAACACACCGTGCATTGGTTCCGGAAAGGATTGCGAATCCACGATAACCCCGCCCTTCGAGAGGGCATTATAGACGCTGTCACATTCCGATGCGTCTTTATAATAGATCCATGGTTCGCCAGCTCTTCAAACGTTGGCATAAACAAATGGAGATTCCTATTGCAATGTTTAGAGGATTTAGATAAAAGTTTAAAGAAGCTTAATTCGAGGCTATTCGTTGTTCGCGGTCAGCCCGCGGATGCGTTGCCAAAATTGTTTCGAGAATGGGGTACAACAGCATTAACCTTTGAAGAGGACCCTGAGCCTTACGGCCGCGTACGCGATCACAATATTATATCTAAATGTCGTGAAGTTGGCATAACAGTGACCTCTCGCGTCTCACACACTTTATACAAATTGGACAAAATAATAGAACGCAATGGTGGTAAAGCTCCGTTAACATATCATCAGTTCCAAGCGTTAATCGCCAGTATGCCGCCCCCTCCACCGGCCGAAGTTACAATAACTCCGCAAATGCTCAATGGAGCCACTACACCGATTACTGATAATCACGATGATCGTTTTGGAGTCCCCACATTAGAAGAGCTTGGATTCGAAACCGAGGGACTAAAGCCACCAATATGGATAGGCGGAGAAAGTGAAGCTTTAGCCAGGTTAGAGAGGCATTTAGAGAGAAAAGCCTGGGTGGCTTCATTTGGAAGACCCAAAATGACACCGCAATCATTGCTGGCGAGCCAAACGGGACTATCACCATACTTAAGATTCGGATGTCTGTCGACCCGATTGTTTTACTATCAATTGACGGAATTGTATAAAAGAGTGAAACGGGTGCGGCCTCCGCTCTCTCTTCACGGACAAATATTATGGAGGGAGTTCTTTTACTGTGCTGCGACAAGAAACCCAAATTTCGATCGCATGGAAGGAAATCCTATATGCGTTCAAATACCTTGGGAGAAGAATCAGGACGCTCTAGCGAAATGGGCAAATGGGCAAACCGGATACCCTTGGATTGATGCCATAATGATTCAATTACGAGAAGAGGGCTGGATTCACCATTTATCGCGACACGCTGTAGCCTGCTTTTTGACAAGAGGAGATCTTTGGATTTCATGGGAAGAAGGAATGAAGGTGTTCGATGAGTTACTCCTCGATGCAGACTGGTCGGTGAACGCAGGCATGTGGATGTGGTTCTCCTGCTCGTCGTTTTTCCAACAATTTTTCCACTGTTACTGCCCTGTGCGTTTTGGAAGGAAAACCGACCCTAATGGTGATTTCATAAGGAAATACATACCGGCTCTGAAGAACATGCCAACGCGCTACATACACGAGCCGTGGATGGCCCCGGAGTCTGTGCAGGCGGCCGCCCAGTGCAGCATCGGACGTGACTACCCCATGCCGATGGTCGACCACACCAAGGCCTCGCAGATAAATATCGAACGGATCAAACAGGTCTACGCACAACTCGCCCGATATAAGCCTCAAGCTACGCTAAATGCTAATGCAGTCCAGCGACCCAACGTGTTGCAGTCGTCGCCAAGTCCGACATCAATAATCGCCAGCATCAACCAGTCCAACTTCTTGTGCAGCCAAACTCCGGATCCACAAAACGCTACATCGCATTCGTTCAAAGAGCCGTCAGACACATTCCCAATAAACAAAAAAAACAAAGATACCGAATCGAACCAAACTCGCTACAAACGAGTAATTATCGTGCAAAAAACACGCAATTCTAATGTGGAAATTAATGTATCCGCTCCACAAGATCGGAGAGCTTCAGATAATTATATCACGATTAATGAGGGAGATCCTGCACACAGTACAGACGCAAAAAAACAAGAAAATAATTACGATTTCAAAAATTTGGCTATCGACAATCATATCCGAGGGTTTAATAACATTCAAAATATAAATTCTCAATATTATTTAACTAATTATTCTACTAAGAAGAAAGAAACTGAAATAAAAAATGAAACAGACGAAGGTCCATTTGTGCGCCCGGAAAGTTATGGCACAGACAGGGCTTACACAATCAACCGTTGCCCTTCCATGTCAAATGAAAAAGAATTTGAAACGCCCAAAGAAAATCTAAAACAATAA
Protein
MSAAPETLPPPSAQAHTPARPTHMSAPRRTPGKHTVHWFRKGLRIHDNPALREGIIDAVTFRCVFIIDPWFASSSNVGINKWRFLLQCLEDLDKSLKKLNSRLFVVRGQPADALPKLFREWGTTALTFEEDPEPYGRVRDHNIISKCREVGITVTSRVSHTLYKLDKIIERNGGKAPLTYHQFQALIASMPPPPPAEVTITPQMLNGATTPITDNHDDRFGVPTLEELGFETEGLKPPIWIGGESEALARLERHLERKAWVASFGRPKMTPQSLLASQTGLSPYLRFGCLSTRLFYYQLTELYKRVKRVRPPLSLHGQILWREFFYCAATRNPNFDRMEGNPICVQIPWEKNQDALAKWANGQTGYPWIDAIMIQLREEGWIHHLSRHAVACFLTRGDLWISWEEGMKVFDELLLDADWSVNAGMWMWFSCSSFFQQFFHCYCPVRFGRKTDPNGDFIRKYIPALKNMPTRYIHEPWMAPESVQAAAQCSIGRDYPMPMVDHTKASQINIERIKQVYAQLARYKPQATLNANAVQRPNVLQSSPSPTSIIASINQSNFLCSQTPDPQNATSHSFKEPSDTFPINKKNKDTESNQTRYKRVIIVQKTRNSNVEINVSAPQDRRASDNYITINEGDPAHSTDAKKQENNYDFKNLAIDNHIRGFNNIQNINSQYYLTNYSTKKKETEIKNETDEGPFVRPESYGTDRAYTINRCPSMSNEKEFETPKENLKQ
Summary
Uniprot
EMBL
HM747058
HM747060
ADM86933.1
BABH01002552
JQ713142
AGA11668.1
+ More
JQ713141 AGA11667.1 NWSH01000412 PCG76580.1 EF117812 ABO38435.1 JQ713140 AGA11666.1 ODYU01012739 SOQ59201.1 JX077109 AFR54427.1 KY491539 AUG44606.1 GQ896503 ADN94465.2 JQ616847 AFJ22639.1 EF396286 ABQ84983.1 KQ460615 KPJ13581.1 KF977409 AHL69753.1 RSAL01000187 RVE44783.1 DQ184682 ABA62409.1 AGBW02009451 OWR50781.1 KQ459602 KPI93335.1 GGFK01000791 MBW34112.1 GGFK01000854 MBW34175.1 GGFK01000797 MBW34118.1 NEVH01008214 PNF34525.1 EF110521 ABO31112.1 ADMH02002109 ETN58942.1 KQ414583 KOC70762.1 UFQS01000097 UFQT01000097 SSW99415.1 SSX19795.1 LC320666 BBD05666.1 KX602318 APP94027.1 GEDC01018842 GEDC01015027 GEDC01008790 JAS18456.1 JAS22271.1 JAS28508.1 JQ713137 AGA11663.1 JQ713136 AGA11662.1
JQ713141 AGA11667.1 NWSH01000412 PCG76580.1 EF117812 ABO38435.1 JQ713140 AGA11666.1 ODYU01012739 SOQ59201.1 JX077109 AFR54427.1 KY491539 AUG44606.1 GQ896503 ADN94465.2 JQ616847 AFJ22639.1 EF396286 ABQ84983.1 KQ460615 KPJ13581.1 KF977409 AHL69753.1 RSAL01000187 RVE44783.1 DQ184682 ABA62409.1 AGBW02009451 OWR50781.1 KQ459602 KPI93335.1 GGFK01000791 MBW34112.1 GGFK01000854 MBW34175.1 GGFK01000797 MBW34118.1 NEVH01008214 PNF34525.1 EF110521 ABO31112.1 ADMH02002109 ETN58942.1 KQ414583 KOC70762.1 UFQS01000097 UFQT01000097 SSW99415.1 SSX19795.1 LC320666 BBD05666.1 KX602318 APP94027.1 GEDC01018842 GEDC01015027 GEDC01008790 JAS18456.1 JAS22271.1 JAS28508.1 JQ713137 AGA11663.1 JQ713136 AGA11662.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
4K0R
E-value=0,
Score=1846
Ontologies
GO
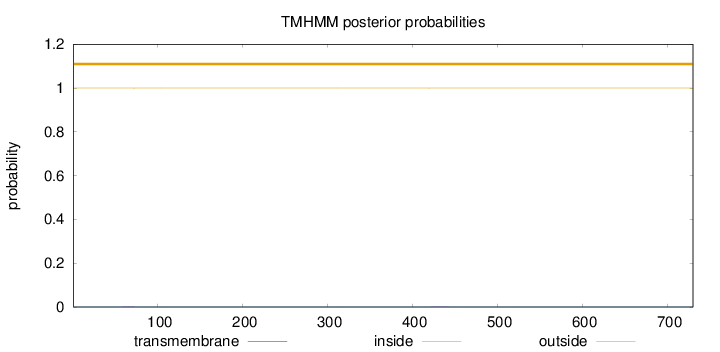
Topology
Length:
730
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00982
Exp number, first 60 AAs:
0.00084
Total prob of N-in:
0.00017
outside
1 - 730
Population Genetic Test Statistics
Pi
222.133066
Theta
192.905104
Tajima's D
0.448241
CLR
0.44693
CSRT
0.496025198740063
Interpretation
Possibly Positive selection
