Pre Gene Modal
BGIBMGA007740
Annotation
PREDICTED:_golgin_subfamily_A_member_4-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.446
Sequence
CDS
ATGTTTAAGAAATTGAAAGATAAACTTGCTGAGGAAGTGAAATCCTCACCACAGAGAATTCAGCAATTCGCCCAAGCCGCCCAGGCTGCTGTAACATCAGCATCAAGTAGTATTTCAGATATCACAAATAGTGACCTATTTTCTATAGGAGAAAATGATAGTCAAACATTTCATTCAACTAAATCACAGCCATCAACTGTCTCTCAAAGTAATACTTTTCGAGATGTCTCGCCCATGCAAACAACAACAACTGAATCAACTGAATACAATTCTGGACAAGACTTAAGTCAAGATAATGAGCGTCAAAGGAGACTGTCTAACAGTTCTTTTGCGAGTGATGTATCCTTCAGACTACCCAGCTACGACACACCAATGATGTACCACTTACAATCCGATATTGAAGTATCTGCAAGTGAAGCTGAAGAAAGAGGCTTAAATGGTAGCACAGTGAGCCTCGATCGTGTCACTAAAGAACAACTGTATGCTGCCTATAAACGAACACAGGAAAGATACAAGAAATATAGGACACAATATGGAGACTTAGCCAGACACTACAAGCTCTTAGAGAGAGAAAATACTAAAGCAAGGAGTGTCTTAGTTGAAACTCAAGATAAAGCCTTACGGAGAATATCGGAATTGAGAGAACAATGCTCACTAGAACAAAGTGCAAAAGCACATTTAGAAAAAGCTCTTCGTGTGGAAATTGAAGAAAAAAATATGAAAATTGAGACTTTGAACACGAAAATACAATTCCTCTCCAATAGTACAAGTGCAGAAATTCCTACCAATATAGTGGATAACAATAAGATTATCATCGACAAAGATAGCAATATACCTTTAATTAATTTAGTAGAAAACGAAACAAAGGTGTCCAATGATGAATCGAAATTGCCAGTGTCATCAGATTTAACAGTGTTAAATGAAAAAATTGAAAAAATGGAATTATTGATACAGAAATATAAAGAATCATTGAAGAGCACAAAAGACAAAAATTCTCAATTAATAACCGAATTGCAAATAATGACTAACAAGCTAGAAAACAAAAGTAAAGAAGTTGAAGAAGTCTCAATACTTAATGAAAAGTACAGAGACGCCAGTGCAAAGATAAAAGAGCTGGAAAATTTAAATGAAGAACTTCAAAATAAAATAAATTCTTATGATTTCAATAAATCGAAAAAAGTTTCTGAACTAGAAACGGATTTACAAAAGGCAAACGAAGAAGTCCAACACCTTCAAAAGAAAATAGAAGTATTTTCTAAGAGAGAAGAGGAATACGCTATTTCTCTAGCAGAGAATAAATTGAGCATCCATAAAGAATTGGAAAGTAAAGAATCCGAAATCAAATCGTTAAAACATAGTTTGACCGCTAGTAAAAAAGAAATACAATCTTTAAGTATTGTCGTCACTGACTATAAAAATAGCATAGAAGCACTCGAACAAGAAAAATCTAAATTAGTTATTGAGAATAGTGAGATAGCAACCTTGAAATCTAAAGTTGAAGAATTAGAAACACATCTACAAGAACTTAATAAAAAAAATCAAATTCAAGAACAACAACACACCAAATTGAATGAAGAATATAAATGCCTGCAGCTTCAATTAAAACAAGAAATGGCTGAAAAATTAGCTATGATGGACAGAAACGCCTATTTAGAAAACCGTATTTCCCAAATATCTGAAGAGAACTCGAAGAAAAGTTCTCAAGTTAATCAATTAGAATCTAAATTACAATCTTTACAAAATGATAAAAGTAACGAAAGCATTGGAAAAGAAAAAGAAATACTGACTGAGCTAAACATTTGGAAGTCCAAGTATGATAAGCTTGAATCAGAGATGCAGGAAGAAAGAGAAGAATTAGTTAAATTACAATCGGAAATAGAAAAGCTGATTGAAAATCACGAACATATTCAAAAACAAAATGTAGAATTTCGGACAACTATCTCGGATTTACAGGCTGAAACTTCAATTTTAAATCGACAACTAAGTCATTATAATAAAATAAAAAACGATACAAATAAAATATTGGCTGATGTGGGAAAACTACGCGAATTAATAAAGTTAACTTCGAAAGATATTTTCGCGAATGTAAAAGACCATGTGGATATGATTAAAACAATTAAAATAGACATTAACAATAATATAAAAGAACTTTCCAACAACGAGCATATCAATTATGTTAATAATGAGGAACTCCGAAATGAGTGTTTGTTACTTCAAAGAAACATAAATGACATAACAGCACTGCAATCTGAAACGAATAAAACTCAAACAAAATTAAAAGAAGAAAATGTCTCTTTAACTTTAACAATACAAAAACTTACTGCAGATTTACAGGAAAAAATTGATGTCGATGAAAAATATAGGAACGCGTCCAATGATTTAAATTCATTAAGAAGTGAAAATCAAAGACTCAGTGTACTTATAAAAGATTATGAAAATACTATACAAAATTTGCGTAATTCATTAGATTTACAAAAATCGCAGTTGTCCACACTACATGAAGAGGTAATTTGCCTTCGAAAAGAAAAAGAGCTCTTTGAAGAAAACTTAAACAAATTGCAGAATATTAACAAAGATTTGGCGTCAGAACAAGACAAACATAAAAACGCAATTGCAACAAATGTAGAAATGATTAATATTTTAAGAACTAAAAATGATGACTTGCTGGAAAAAAGCGATGAATTGAACAATAAGTTAATAAACTACCAACAACTAGAAGAACATCATAAGTTATTAACACAAGAAAACTCTTTTCTTAAAAGTCAAACTGAAAATATCGCAAGCAATGTACGAGAATTAGAATTCGAAATAAATGAAGTAAGAAAATCTCACTCTGCTATTGAATCAGAAAAAGAACATTTAAGTAATGTCATCGAACAAATAGAAAACCGACGTACATTGCTAGTTTCAAAAGATACACAAACAGAATACAACGAAATAGAAACGGAACTCGACAAAGAAAAAAACTTATCTAATATAAAGATAATAAATGAACTTAGCGAGAAGTATGATGCCTTAAAAGGAGAAAATAGACGGCTTTTGTCTGACATTGAAGGACTCCAAACACATTTAACGAAAACATCAAAAGAAAACAGCTTACTCAATGACAAACTCCGTGAGCAAATTGCAACAAGTGAAACAGGGAACTACATGTCCCATGATTTGAATAATGACATTCGGTTGGCTAAAGATAAAATTGATCACTTAACAAGGGAGAATACACTTTTAAATGAAGAAAACCTGGAACTAAAAGACCAGTTACAGTCACAGGAATATTGCAAACCTACAAATGCTGATCAGTGTTTTGAACACAAAGGGGAAAATGAGCTGTCTGAAATTAAAGAAAAATATTACACATTAATACTGGCTAATAGCAAAATAGAACAGAAAAACATTGATTTGGAACAATTGAACAGATCATTGAATGTTAACATCCAAGATGTACAAGCCAAAAACGAAAAATTGAAACTTTCGAATGAAAAAATAGAGCGCAGGCTCGATGAAGCTTTAGTTAGTTTAAGACATTTGCACTCTTTAGAAGAAAATACCGAGCTAGAATATTTAAGGAATATACTCTATGAATACCTTACAGGCTCAGGAACACATTCCATTACTTTAGCGAAAGTTTTAGCTGCAGTAGTCAAATTTGATGACAAGCAAACCCAAATGGTTCTTCAAAAAGAAAAAGAAAGACAAGGTTTTCTAAGGCAACTCCGACTCACGTGA
Protein
MFKKLKDKLAEEVKSSPQRIQQFAQAAQAAVTSASSSISDITNSDLFSIGENDSQTFHSTKSQPSTVSQSNTFRDVSPMQTTTTESTEYNSGQDLSQDNERQRRLSNSSFASDVSFRLPSYDTPMMYHLQSDIEVSASEAEERGLNGSTVSLDRVTKEQLYAAYKRTQERYKKYRTQYGDLARHYKLLERENTKARSVLVETQDKALRRISELREQCSLEQSAKAHLEKALRVEIEEKNMKIETLNTKIQFLSNSTSAEIPTNIVDNNKIIIDKDSNIPLINLVENETKVSNDESKLPVSSDLTVLNEKIEKMELLIQKYKESLKSTKDKNSQLITELQIMTNKLENKSKEVEEVSILNEKYRDASAKIKELENLNEELQNKINSYDFNKSKKVSELETDLQKANEEVQHLQKKIEVFSKREEEYAISLAENKLSIHKELESKESEIKSLKHSLTASKKEIQSLSIVVTDYKNSIEALEQEKSKLVIENSEIATLKSKVEELETHLQELNKKNQIQEQQHTKLNEEYKCLQLQLKQEMAEKLAMMDRNAYLENRISQISEENSKKSSQVNQLESKLQSLQNDKSNESIGKEKEILTELNIWKSKYDKLESEMQEEREELVKLQSEIEKLIENHEHIQKQNVEFRTTISDLQAETSILNRQLSHYNKIKNDTNKILADVGKLRELIKLTSKDIFANVKDHVDMIKTIKIDINNNIKELSNNEHINYVNNEELRNECLLLQRNINDITALQSETNKTQTKLKEENVSLTLTIQKLTADLQEKIDVDEKYRNASNDLNSLRSENQRLSVLIKDYENTIQNLRNSLDLQKSQLSTLHEEVICLRKEKELFEENLNKLQNINKDLASEQDKHKNAIATNVEMINILRTKNDDLLEKSDELNNKLINYQQLEEHHKLLTQENSFLKSQTENIASNVRELEFEINEVRKSHSAIESEKEHLSNVIEQIENRRTLLVSKDTQTEYNEIETELDKEKNLSNIKIINELSEKYDALKGENRRLLSDIEGLQTHLTKTSKENSLLNDKLREQIATSETGNYMSHDLNNDIRLAKDKIDHLTRENTLLNEENLELKDQLQSQEYCKPTNADQCFEHKGENELSEIKEKYYTLILANSKIEQKNIDLEQLNRSLNVNIQDVQAKNEKLKLSNEKIERRLDEALVSLRHLHSLEENTELEYLRNILYEYLTGSGTHSITLAKVLAAVVKFDDKQTQMVLQKEKERQGFLRQLRLT
Summary
EMBL
Proteomes
Pfam
PF01465 GRIP
Interpro
IPR000237
GRIP_dom
ProteinModelPortal
PDB
1UPT
E-value=0.000205636,
Score=111
Ontologies
GO
Topology
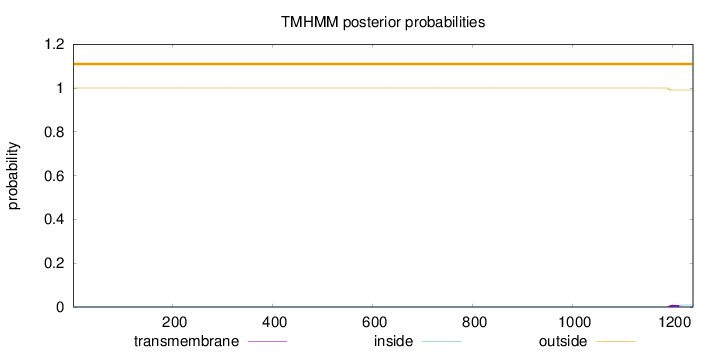
Length:
1241
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19297
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00001
outside
1 - 1241
Population Genetic Test Statistics
Pi
161.808927
Theta
156.944589
Tajima's D
-0.838211
CLR
18.771628
CSRT
0.171591420428979
Interpretation
Uncertain
