Gene
KWMTBOMO08969
Pre Gene Modal
BGIBMGA007792
Annotation
ABC_transporter_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.819
Sequence
CDS
ATGAATAGTGATGGGAGAGCCGGAGAAAATTCTAGTGCTGAGAAGCGAAGGAAGCCCCATAAGCCGAACATTTTGTCTCGTATATTTCTATGGTGGATGTGCCCCGTACTCGTTAAAGGAAATCAAAGAGACATTGTCGAGGATGACCTCATAATTCCGAAAAAGTCATTTAATTCTGAAAATCAAGGAGAGTATTTAGAAAGGTATTGGCTCCAAGAATATGAAGCCGCGATAAAGGAGAAAAGGGAGCCGTCATTGTGGACAGCTTTACGAAAAGCTTACTGGTTGGGATACATGCCGGGAGCTATCTACTTGATAATCATATCTGTCTTCAGGATAATTCAGCCGCTAGTATTCGCCGAACTGCTTTCCTACTGGTCAGTAGAAGCAACTATAACTCGACTAGAAGCAAGCTATTATGCTCTAGCACTTTTAGGTATAAACTTCATTAATATGATGTGTCAACATCACAATTCATTGTTTGTAGCGCGCTTTGGACTCAAGGTTAAAGTGGCTTGCTCATCTTTAGTATATAGAAAGTTGTTGCGCATGGACCAAGTAGCCTTAGGAGATGTAAGCGGAGGGAAACTGGTGAACCTGTTATCTAATGATGTTGCTAGATTCGACTACGCATTCATGTTTTTGCACTACCTCTGGGTGGTACCAGTTCAAGCGGCTGTAGTATTATATTTCTTATATTATATTTCTGCGGGATACGCACCGTTCGTCGGCTTCTTTGGGGTTGTGATTTTAATTCTACCTATACAAGCTGGTCTCACAAAATTGACCTCTGTGGTCAGAAGAGAAACTGCTCAGAGAACCGACCGAAGGATAAAATTGATGACGGAAATTATTAATGGCATTCAGGTTATTAAAATGTACGCATGGGAGAAACCGTTCCAAGCAATCGTCAAGGTGGCAAGAAACTTCGAAATGATAGCACTCAGGAAGTCTATATTCATTCGGAGTGTTTTCCTTGGTTTCATGCTGTTCACTGAAAGAAGTATAATATTTATAACATGTTTGACATTGTTACTTACGGGTAACCTTGTTACAGCTACTCTGATTTACCCTATTCAACAATACATCAGCATTATTCAAATCAATTTGACTATGATACTGCCTCTTGCCATTGCAAGTTTGTCTGAAATGCTGGTCTCGTTGGAACGCATTCAAAATTTCTTGGTGAAAGATGAAAGAGAGGACGTCCAAATAACACCAAAATCTTATGGAGATGATAATAGATTGATTTTCAACAACAAGGCATCGCTTGGCCCTCAAAACGAAATTATCCCAAAGAAATATTTAGCCACCGATGGCCAACTTGCCTCAACATTAACGAATGAACCAGTGCTTCAAACTGACCCAGCCGTGTGTGACTATCCCATAGAATTATCTAAAGTAGACGCCACATGGTCGTCATCAACAGATACTTCAGAAATGACACTAAGGAATATTAGTCTCAGAATAGGCAGAGGCAAACTTTGTGCTATAATTGGACCTGTTGGATCAGGAAAGTCATCCATTTTGCAAGTTCTTCTGAAAGAACTGCCGGTTTGCGGTGGTAGCCTGAGAATAAATGGGAGGCTTTCCTACGCGTGTCAAGAGTCTTGGCTGTTCCCTGCTACTGTGAGAGAAAACATTTTGTTTGGTTTGCCGTACGAATCACAGAAATATCACGAGGTGTGTAAAGCTTGTTCGTTGCTTCCTGATTTCAAACAATTTCCGTACGGAGACCTCTCCTTAGTTGGAGAGCGAGGAGTATCCTTGTCAGGGGGACAGCGTGCCAGAATTAATCTCGCTAGGGCTGTTTACCGAGAGGCCGACATTTACTTGCTAGATGATCCTCTCTCAGCAGTGGACGCCAACGTTGGCAGGCAGCTGTTTGACGGCTGTATCAAAGGATATCTACGGGGTCGAACTTGTGTCCTTGTAACCCATCAAATCCACTATCTTAAAGCAGCTGACATTATTGTCATACTGAACGAGGGGGCAATTGAAAACGTTGGTAGCTACGATGATTTGGTCAAGACCGGAACAGAGTTTTCGAAGCTGTTGACTAATCAAGAAAGCAACGATAATGAGAATGGCCCGGAAAAGAATTTCTTACGGGCTATATCGAAAATATCTACTAAGAGTGTTGAAGATCCAGATAACGAGAAGGTACAAGTGGAGGAAGAAGAGAAACGTGCCAAAGGAAATTTGAAATTCTCTGTTTTATACAAGTATTTGTCTGCAGTGAAATCATGGTTTCTAGTATTTTTGATGGTAGCCACATTAGTTATCACACAAGGATGTGCGATGTTCATAGATTATTGGCTGAGTTTTTGGACAAATCAAGTTGATGAGTATGAACAATCTTTAGCTGAGGGAGAAGAACCGAGCACAAGTTTGGACACGCAAGCCGGAGCGTATACTTTAGGAGTATACCTTTGGACTTATGGCGGAGTCATCTTAATCCTCATAGTGATCAGTCACGTCAGAATTTTAACTTTCGTCATTACGACTATGAGAGCTTCATCTAATTTTCATGATACCGTTTACAAAAAGTTGATTATCACAGTTATGCGATTCTTTGACATGAATCCCTCAGGTCGTGTGTTGAATCGTTTTTCAAAGGATATGGGTGCGATGGACGAATTCCTTCCGAGGAGTTTATTTGAAACTGTTCAGATGTATTTAACATTATGCAGTATACTTATTTTAAATGCTATAGCATTACCGTGGACGTTGATCCCAACAGCGGTACTTTTGATTCTCTTCTTCTTTCTCCTGAAATGGTACTTAAATGCTGCTCAAGCTGTAAAAAGATTAGAAGGCACAACTAAGAGTCCAGTACTCGGCATGATCAACTCTACCCTCACAGGTCTGTCTACGATCCGAAGTTCTAATTCACAAGGACGACTGCTAGAAATGTTTGATAACGCACAGAATCTCCACACGAGTGCATTTTATACGTTTGTTGGTGGATCAACTGCATTTGGTTTATATCTGGATGCGCTTTGCTTAGTTTATTTGGGAGTGATATTGACGATCTTCCTTGTAATCGACTTCAGTACTTTAATTCCCGTCGGTAGCGTGGGCTTGGCTGTGAGTCAGTCTATGGTGTTAACGATGATGCTGCAAATGGCGGCTCGATTCACAGCCGACTTCCTCGGACAAATGACGGCTGTCGAGAGAGTTCTCGAATATACCGAGCTTCCGATGGAAGAAAACATGTATGATGGATCCCAACTTCCCAAGGACTGGCCTACACACGGTCATATTGAATTTCAAAATCTATTTTTGAACTATTCTCAAGAAGATCCACCAGTACTCAAAGATTTAAACTTTGTAATTGAGAATGGTTGGAAGGTTGGAGTAGTCGGTCGAACCGGTGCCGGCAAGTCTTCTATGATATCCGCACTGTTCAGGCTGTACGACCTGCAAGGACACATCAGAATAGACGGAATCGATACCAACATCATCGCTAAAACGGAGCTAAGATCGAAGATTTCTATCATACCACAAGAACCGATTTTGTTCTCTGCATCGGTGCGATACAATTTGGACCCCTTCGACAGTTATAGCGACGACGAAATTTGGAGAGCTTTGGAACAGGTTGAACTGAAAGAGGTAATTCCAGCGCTCGATTATAAAGTGTCAGAAGGCGGCAGCAACTTTTCCGTCGGTCAGCGACAACTGGTATGTTTGGCTCGAGCTGTTTTGCGTTCTAACAAAATCCTTGTAATGGACGAGGCGACAGCCAACGTTGATCCTCAAACTGACGCGTTGATTCAGACGACGATACGTCGCGAGTTTGCGTCATGTACGGTGATCACGATCGCGCACCGACTCAACACGATCATGGATTCTGATCGCGTCCTGGTCATGGACAAAGGAGTTGTTGCCGAATACGATACTCCTTATGCTCTGCTCTCCGACCCCAATTCGATCTTCAGCTCGATGGTGCGCGAAACAGGAGATACAATGAGCAAAGTCCTCTTCCGAGTGGCTGAAGATAAACATCTTGGTAGAAATACAGAAAAATGA
Protein
MNSDGRAGENSSAEKRRKPHKPNILSRIFLWWMCPVLVKGNQRDIVEDDLIIPKKSFNSENQGEYLERYWLQEYEAAIKEKREPSLWTALRKAYWLGYMPGAIYLIIISVFRIIQPLVFAELLSYWSVEATITRLEASYYALALLGINFINMMCQHHNSLFVARFGLKVKVACSSLVYRKLLRMDQVALGDVSGGKLVNLLSNDVARFDYAFMFLHYLWVVPVQAAVVLYFLYYISAGYAPFVGFFGVVILILPIQAGLTKLTSVVRRETAQRTDRRIKLMTEIINGIQVIKMYAWEKPFQAIVKVARNFEMIALRKSIFIRSVFLGFMLFTERSIIFITCLTLLLTGNLVTATLIYPIQQYISIIQINLTMILPLAIASLSEMLVSLERIQNFLVKDEREDVQITPKSYGDDNRLIFNNKASLGPQNEIIPKKYLATDGQLASTLTNEPVLQTDPAVCDYPIELSKVDATWSSSTDTSEMTLRNISLRIGRGKLCAIIGPVGSGKSSILQVLLKELPVCGGSLRINGRLSYACQESWLFPATVRENILFGLPYESQKYHEVCKACSLLPDFKQFPYGDLSLVGERGVSLSGGQRARINLARAVYREADIYLLDDPLSAVDANVGRQLFDGCIKGYLRGRTCVLVTHQIHYLKAADIIVILNEGAIENVGSYDDLVKTGTEFSKLLTNQESNDNENGPEKNFLRAISKISTKSVEDPDNEKVQVEEEEKRAKGNLKFSVLYKYLSAVKSWFLVFLMVATLVITQGCAMFIDYWLSFWTNQVDEYEQSLAEGEEPSTSLDTQAGAYTLGVYLWTYGGVILILIVISHVRILTFVITTMRASSNFHDTVYKKLIITVMRFFDMNPSGRVLNRFSKDMGAMDEFLPRSLFETVQMYLTLCSILILNAIALPWTLIPTAVLLILFFFLLKWYLNAAQAVKRLEGTTKSPVLGMINSTLTGLSTIRSSNSQGRLLEMFDNAQNLHTSAFYTFVGGSTAFGLYLDALCLVYLGVILTIFLVIDFSTLIPVGSVGLAVSQSMVLTMMLQMAARFTADFLGQMTAVERVLEYTELPMEENMYDGSQLPKDWPTHGHIEFQNLFLNYSQEDPPVLKDLNFVIENGWKVGVVGRTGAGKSSMISALFRLYDLQGHIRIDGIDTNIIAKTELRSKISIIPQEPILFSASVRYNLDPFDSYSDDEIWRALEQVELKEVIPALDYKVSEGGSNFSVGQRQLVCLARAVLRSNKILVMDEATANVDPQTDALIQTTIRREFASCTVITIAHRLNTIMDSDRVLVMDKGVVAEYDTPYALLSDPNSIFSSMVRETGDTMSKVLFRVAEDKHLGRNTEK
Summary
Uniprot
G1UHW7
G1UHW8
I1VH51
A0A0K1SB04
A0A0K1SB50
D8V116
+ More
A0A2A4JM73 A0A2H1WJQ9 A0A1Z2RPL3 A0A2H4YHT1 A0A2H4YHT5 A0A2K9P5A5 A0A2H4YHR9 D8V119 A0A2H4YHT2 A0A2H4YHR7 A0A2H4YHT7 A0A0B4ZS88 A0A088C848 A0A2L1K5C6 A0A060D135 A0A1B0RHN5 A0A3G1PY78 A0A1B0RHP5 A0A1N7TA09 A0A2H4YHV2 A0A2H4YHS1 A0A2H4YHT4 A0A2H4YHU4 A0A0B4ZX17 R9WPG7 G3JWY5 A0A0E3ZDK3 A0A097DCY9 A0A0B4ZYY9 A0A3S2LNC5 G3JWY1 A0A194PJX3 G3JWY2 A0A212FDD7 A0A223PQG6 A0A060D4Q3 A0A0F6PPP5 A0A088C894 A0A0F6PQI8 A0A3G1PY80 A0A2K9P5C0 A0A060DA23 A0A060DAW3 A0A212FDE8 A0A2A4JLC9 A0A0B4ZWR3 A0A0E3ZCM1 A0A194R7E2 A0A3S2PG34 A0A223PQF2 A0A194PJ23 A0A0B4ZTU5 G1UHY0 A0A0E3UUF7 A0A3G1PY98 A0A3S2TFR5 A0A2H1X0D3 A0A2H1WJI9 D6WK76 A0A2H4KAX0 A0A146LDV6 A0A0C9PVS6 A0A2M4B9I8 A0A3R5SMP1 A0A2A4K8A3 A0A158V0F7 A0A182FEZ4 A0A0P4W3Q5 A0A0V0G5R1 K7IU38 A0A3L8DIP4 A0A1Y1N3T5 F4X6C1 A0A1Y1MZC5 A0A151ILB5 A0A1I8NI62 W5JPP8 A0A0L0CKR5 A0A1I8Q0X0 A0A2A3EMT6 B4JUQ0 A0A1L8DUP6 Q7Q5I0 A0A1L8DV41 A0A0L0CGT9 A0A3L8DIZ7 A0A1S4GS60 A0A158NLW9 A0A1I8NAJ3 A0A182RML1 T1PE30 A0A1I8Q3P7 A0A1J1ITZ7 A0A1Q3F1A4
A0A2A4JM73 A0A2H1WJQ9 A0A1Z2RPL3 A0A2H4YHT1 A0A2H4YHT5 A0A2K9P5A5 A0A2H4YHR9 D8V119 A0A2H4YHT2 A0A2H4YHR7 A0A2H4YHT7 A0A0B4ZS88 A0A088C848 A0A2L1K5C6 A0A060D135 A0A1B0RHN5 A0A3G1PY78 A0A1B0RHP5 A0A1N7TA09 A0A2H4YHV2 A0A2H4YHS1 A0A2H4YHT4 A0A2H4YHU4 A0A0B4ZX17 R9WPG7 G3JWY5 A0A0E3ZDK3 A0A097DCY9 A0A0B4ZYY9 A0A3S2LNC5 G3JWY1 A0A194PJX3 G3JWY2 A0A212FDD7 A0A223PQG6 A0A060D4Q3 A0A0F6PPP5 A0A088C894 A0A0F6PQI8 A0A3G1PY80 A0A2K9P5C0 A0A060DA23 A0A060DAW3 A0A212FDE8 A0A2A4JLC9 A0A0B4ZWR3 A0A0E3ZCM1 A0A194R7E2 A0A3S2PG34 A0A223PQF2 A0A194PJ23 A0A0B4ZTU5 G1UHY0 A0A0E3UUF7 A0A3G1PY98 A0A3S2TFR5 A0A2H1X0D3 A0A2H1WJI9 D6WK76 A0A2H4KAX0 A0A146LDV6 A0A0C9PVS6 A0A2M4B9I8 A0A3R5SMP1 A0A2A4K8A3 A0A158V0F7 A0A182FEZ4 A0A0P4W3Q5 A0A0V0G5R1 K7IU38 A0A3L8DIP4 A0A1Y1N3T5 F4X6C1 A0A1Y1MZC5 A0A151ILB5 A0A1I8NI62 W5JPP8 A0A0L0CKR5 A0A1I8Q0X0 A0A2A3EMT6 B4JUQ0 A0A1L8DUP6 Q7Q5I0 A0A1L8DV41 A0A0L0CGT9 A0A3L8DIZ7 A0A1S4GS60 A0A158NLW9 A0A1I8NAJ3 A0A182RML1 T1PE30 A0A1I8Q3P7 A0A1J1ITZ7 A0A1Q3F1A4
Pubmed
EMBL
AB620074
BAK82126.1
AB620075
BAK82127.1
JQ774504
AFI44049.1
+ More
KP400523 AKV71230.1 KP400524 AKV71231.1 GQ332571 ADH16740.1 NWSH01001057 PCG72886.1 ODYU01009083 SOQ53247.1 KY489760 ASA45739.1 MG387056 AUE23770.1 MG387055 AUE23769.1 KY646296 AUO38740.1 MG387043 AUE23766.1 GQ332573 ADH16744.1 MG387057 AUE23771.1 MG387051 AUE23768.1 MG387058 AUE23772.1 KJ930171 AJD79136.1 KF479231 AHL68986.1 MF929077 AVE17269.1 KF926099 AIB06821.1 KM360184 AKH49600.1 KY701524 AVV62499.1 KM360192 AKH49608.1 KM068116 AJY53612.1 MG387063 AUE23774.1 MG387050 AUE23767.1 MG387068 AUE23775.1 MG387062 AUE23773.1 KJ930167 AJD79134.1 KC758896 AGO01048.1 JN030495 AEI27596.1 KM245561 AKC96180.1 KM103521 AIS93186.1 KJ930169 AJD79135.1 RSAL01000049 RVE50388.1 JN030490 AEI27592.1 KQ459602 KPI93323.1 JN030491 AEI27593.1 AGBW02009058 OWR51775.1 KX710207 ASU47347.1 KF926100 AIB06822.1 KM232756 AKC34055.1 KF479232 AHL68987.1 KM250380 AKC34056.1 AVV62500.1 KY646297 AUO38741.1 KF926101 AIB06823.1 KF926102 AIB06824.1 OWR51776.1 PCG72885.1 KJ930170 AJD79133.1 KM245562 AKC96181.1 KQ460615 KPJ13567.1 RVE50389.1 KX710208 ASU47346.1 KPI93322.1 KJ930168 AJD79132.1 AB621548 BAK86406.1 KM245560 AKC96179.1 AVV62497.1 RSAL01000187 RVE44791.1 ODYU01012016 SOQ58094.1 SOQ53245.1 KQ971342 EFA03948.2 KY484779 ASS36021.1 GDHC01020387 GDHC01013422 JAP98241.1 JAQ05207.1 GBYB01005573 JAG75340.1 GGFJ01000531 MBW49672.1 MH172501 QAA95908.1 NWSH01000076 PCG79892.1 KF828786 AIN44103.1 GDKW01000545 JAI56050.1 GECL01002731 JAP03393.1 AAZX01002657 QOIP01000007 RLU20304.1 GEZM01016983 GEZM01016981 JAV90996.1 GL888807 EGI58005.1 GEZM01016982 JAV90994.1 KQ977121 KYN05546.1 ADMH02000892 ETN64739.1 JRES01000264 KNC32832.1 KZ288215 PBC32516.1 CH916374 EDV91220.1 GFDF01003930 JAV10154.1 AAAB01008960 EAA10720.3 GFDF01003929 JAV10155.1 JRES01000405 KNC31638.1 RLU20387.1 ADTU01019789 ADTU01019790 KA646208 AFP60837.1 CVRI01000059 CRL03663.1 GFDL01013712 JAV21333.1
KP400523 AKV71230.1 KP400524 AKV71231.1 GQ332571 ADH16740.1 NWSH01001057 PCG72886.1 ODYU01009083 SOQ53247.1 KY489760 ASA45739.1 MG387056 AUE23770.1 MG387055 AUE23769.1 KY646296 AUO38740.1 MG387043 AUE23766.1 GQ332573 ADH16744.1 MG387057 AUE23771.1 MG387051 AUE23768.1 MG387058 AUE23772.1 KJ930171 AJD79136.1 KF479231 AHL68986.1 MF929077 AVE17269.1 KF926099 AIB06821.1 KM360184 AKH49600.1 KY701524 AVV62499.1 KM360192 AKH49608.1 KM068116 AJY53612.1 MG387063 AUE23774.1 MG387050 AUE23767.1 MG387068 AUE23775.1 MG387062 AUE23773.1 KJ930167 AJD79134.1 KC758896 AGO01048.1 JN030495 AEI27596.1 KM245561 AKC96180.1 KM103521 AIS93186.1 KJ930169 AJD79135.1 RSAL01000049 RVE50388.1 JN030490 AEI27592.1 KQ459602 KPI93323.1 JN030491 AEI27593.1 AGBW02009058 OWR51775.1 KX710207 ASU47347.1 KF926100 AIB06822.1 KM232756 AKC34055.1 KF479232 AHL68987.1 KM250380 AKC34056.1 AVV62500.1 KY646297 AUO38741.1 KF926101 AIB06823.1 KF926102 AIB06824.1 OWR51776.1 PCG72885.1 KJ930170 AJD79133.1 KM245562 AKC96181.1 KQ460615 KPJ13567.1 RVE50389.1 KX710208 ASU47346.1 KPI93322.1 KJ930168 AJD79132.1 AB621548 BAK86406.1 KM245560 AKC96179.1 AVV62497.1 RSAL01000187 RVE44791.1 ODYU01012016 SOQ58094.1 SOQ53245.1 KQ971342 EFA03948.2 KY484779 ASS36021.1 GDHC01020387 GDHC01013422 JAP98241.1 JAQ05207.1 GBYB01005573 JAG75340.1 GGFJ01000531 MBW49672.1 MH172501 QAA95908.1 NWSH01000076 PCG79892.1 KF828786 AIN44103.1 GDKW01000545 JAI56050.1 GECL01002731 JAP03393.1 AAZX01002657 QOIP01000007 RLU20304.1 GEZM01016983 GEZM01016981 JAV90996.1 GL888807 EGI58005.1 GEZM01016982 JAV90994.1 KQ977121 KYN05546.1 ADMH02000892 ETN64739.1 JRES01000264 KNC32832.1 KZ288215 PBC32516.1 CH916374 EDV91220.1 GFDF01003930 JAV10154.1 AAAB01008960 EAA10720.3 GFDF01003929 JAV10155.1 JRES01000405 KNC31638.1 RLU20387.1 ADTU01019789 ADTU01019790 KA646208 AFP60837.1 CVRI01000059 CRL03663.1 GFDL01013712 JAV21333.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
G1UHW7
G1UHW8
I1VH51
A0A0K1SB04
A0A0K1SB50
D8V116
+ More
A0A2A4JM73 A0A2H1WJQ9 A0A1Z2RPL3 A0A2H4YHT1 A0A2H4YHT5 A0A2K9P5A5 A0A2H4YHR9 D8V119 A0A2H4YHT2 A0A2H4YHR7 A0A2H4YHT7 A0A0B4ZS88 A0A088C848 A0A2L1K5C6 A0A060D135 A0A1B0RHN5 A0A3G1PY78 A0A1B0RHP5 A0A1N7TA09 A0A2H4YHV2 A0A2H4YHS1 A0A2H4YHT4 A0A2H4YHU4 A0A0B4ZX17 R9WPG7 G3JWY5 A0A0E3ZDK3 A0A097DCY9 A0A0B4ZYY9 A0A3S2LNC5 G3JWY1 A0A194PJX3 G3JWY2 A0A212FDD7 A0A223PQG6 A0A060D4Q3 A0A0F6PPP5 A0A088C894 A0A0F6PQI8 A0A3G1PY80 A0A2K9P5C0 A0A060DA23 A0A060DAW3 A0A212FDE8 A0A2A4JLC9 A0A0B4ZWR3 A0A0E3ZCM1 A0A194R7E2 A0A3S2PG34 A0A223PQF2 A0A194PJ23 A0A0B4ZTU5 G1UHY0 A0A0E3UUF7 A0A3G1PY98 A0A3S2TFR5 A0A2H1X0D3 A0A2H1WJI9 D6WK76 A0A2H4KAX0 A0A146LDV6 A0A0C9PVS6 A0A2M4B9I8 A0A3R5SMP1 A0A2A4K8A3 A0A158V0F7 A0A182FEZ4 A0A0P4W3Q5 A0A0V0G5R1 K7IU38 A0A3L8DIP4 A0A1Y1N3T5 F4X6C1 A0A1Y1MZC5 A0A151ILB5 A0A1I8NI62 W5JPP8 A0A0L0CKR5 A0A1I8Q0X0 A0A2A3EMT6 B4JUQ0 A0A1L8DUP6 Q7Q5I0 A0A1L8DV41 A0A0L0CGT9 A0A3L8DIZ7 A0A1S4GS60 A0A158NLW9 A0A1I8NAJ3 A0A182RML1 T1PE30 A0A1I8Q3P7 A0A1J1ITZ7 A0A1Q3F1A4
A0A2A4JM73 A0A2H1WJQ9 A0A1Z2RPL3 A0A2H4YHT1 A0A2H4YHT5 A0A2K9P5A5 A0A2H4YHR9 D8V119 A0A2H4YHT2 A0A2H4YHR7 A0A2H4YHT7 A0A0B4ZS88 A0A088C848 A0A2L1K5C6 A0A060D135 A0A1B0RHN5 A0A3G1PY78 A0A1B0RHP5 A0A1N7TA09 A0A2H4YHV2 A0A2H4YHS1 A0A2H4YHT4 A0A2H4YHU4 A0A0B4ZX17 R9WPG7 G3JWY5 A0A0E3ZDK3 A0A097DCY9 A0A0B4ZYY9 A0A3S2LNC5 G3JWY1 A0A194PJX3 G3JWY2 A0A212FDD7 A0A223PQG6 A0A060D4Q3 A0A0F6PPP5 A0A088C894 A0A0F6PQI8 A0A3G1PY80 A0A2K9P5C0 A0A060DA23 A0A060DAW3 A0A212FDE8 A0A2A4JLC9 A0A0B4ZWR3 A0A0E3ZCM1 A0A194R7E2 A0A3S2PG34 A0A223PQF2 A0A194PJ23 A0A0B4ZTU5 G1UHY0 A0A0E3UUF7 A0A3G1PY98 A0A3S2TFR5 A0A2H1X0D3 A0A2H1WJI9 D6WK76 A0A2H4KAX0 A0A146LDV6 A0A0C9PVS6 A0A2M4B9I8 A0A3R5SMP1 A0A2A4K8A3 A0A158V0F7 A0A182FEZ4 A0A0P4W3Q5 A0A0V0G5R1 K7IU38 A0A3L8DIP4 A0A1Y1N3T5 F4X6C1 A0A1Y1MZC5 A0A151ILB5 A0A1I8NI62 W5JPP8 A0A0L0CKR5 A0A1I8Q0X0 A0A2A3EMT6 B4JUQ0 A0A1L8DUP6 Q7Q5I0 A0A1L8DV41 A0A0L0CGT9 A0A3L8DIZ7 A0A1S4GS60 A0A158NLW9 A0A1I8NAJ3 A0A182RML1 T1PE30 A0A1I8Q3P7 A0A1J1ITZ7 A0A1Q3F1A4
PDB
5UJA
E-value=0,
Score=1644
Ontologies
KEGG
GO
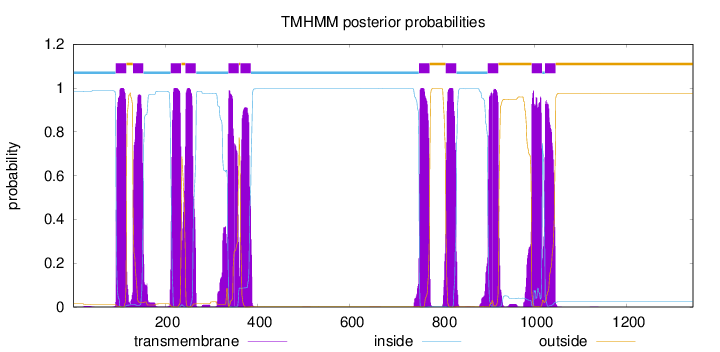
Topology
Length:
1343
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
249.379030000001
Exp number, first 60 AAs:
0.07952
Total prob of N-in:
0.98516
inside
1 - 92
TMhelix
93 - 115
outside
116 - 129
TMhelix
130 - 152
inside
153 - 211
TMhelix
212 - 234
outside
235 - 243
TMhelix
244 - 266
inside
267 - 336
TMhelix
337 - 359
outside
360 - 362
TMhelix
363 - 385
inside
386 - 749
TMhelix
750 - 772
outside
773 - 807
TMhelix
808 - 830
inside
831 - 898
TMhelix
899 - 921
outside
922 - 993
TMhelix
994 - 1016
inside
1017 - 1022
TMhelix
1023 - 1045
outside
1046 - 1343
Population Genetic Test Statistics
Pi
183.412551
Theta
156.088605
Tajima's D
0.601206
CLR
0.034426
CSRT
0.544472776361182
Interpretation
Possibly Positive selection
