Gene
KWMTBOMO08967
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.355 Nuclear Reliability : 1.558
Sequence
CDS
ATGGGCGTTCCCATGCATCTGATTCAGCTGATGAGAGCCCTGTACCTGTCGGGACGGGGATCGGTACGGATCGGCCCAGCACAATCAAGAGAGTTCCGATTTGAGAAAGGAGTGCGCCAGGGTTGCATCGTGTCGCCCATACTTTTTAACATCTATGGCGAATACATAATGCGCAAAACCCTAGAGGAGTGGGATGGTGGCGTCACAGTTGGGGGCGTAAAGATCTCGAACTTACGATACGCTGATGATACCACATTAGTGGCGTCATCAGAAGAGGAAATGGAGGAGCTGCTGCGACGTCTGGTTATAGTTAGCGAGGAAATGGGACTAAAGATCAACCAGTCCAAAACAAAGATCATGATCGTGGATAAATACGGCACCCTTAACGAAAATAATATTCTTGGGCAATACGACATCGTCAAGACTTTCGTGTACCTCGGCTCGATTATTTCCAATAGAGGTGGATCGGAAACCGAAATAAGGCGACGCATCGGAATGGCCAAATCCGCTATGAGCCAGCTAAACAAAATATGGAGAGATCGAAACATACTCAAGAAAACGAAAGTGCAAATTGTTGAGACGCTCGTTTTCTTAATCTTTTTGTATGGGGCTGAGACCTGGATCTTGAAAGAGTCTGACAGGCGACGCATAGACGCGTTTGAGATGTGGTGCTGGCGGCGCATGTTGCGCATCCCTTGGACGGCTTTCCGGACGAACGCTTCCATCCTCAACGAGCTAAAAGTTGACCAGAGGCTATCCACCAAATGTCTATATAGAGTGCTCCAATACTTTGGTCATATAGCAAGAAAACCCTCTGACAACCTGGAGAGACTCATAGTGACGGGTAAAGTGGATGGAAGACGATCCAGAGGTCGTAGCCGAAAAAGATGGAGTGACCAAGTGTCTGAGTGTCTGGACATGAGACTAAGCCACGCACTTCACTGTGCCACAGACCGACATAAGTGGCTACAAATTACAAAAAAAGGTCACAGGCGGGCGCATCACGATGCTCAGCAATGA
Protein
MGVPMHLIQLMRALYLSGRGSVRIGPAQSREFRFEKGVRQGCIVSPILFNIYGEYIMRKTLEEWDGGVTVGGVKISNLRYADDTTLVASSEEEMEELLRRLVIVSEEMGLKINQSKTKIMIVDKYGTLNENNILGQYDIVKTFVYLGSIISNRGGSETEIRRRIGMAKSAMSQLNKIWRDRNILKKTKVQIVETLVFLIFLYGAETWILKESDRRRIDAFEMWCWRRMLRIPWTAFRTNASILNELKVDQRLSTKCLYRVLQYFGHIARKPSDNLERLIVTGKVDGRRSRGRSRKRWSDQVSECLDMRLSHALHCATDRHKWLQITKKGHRRAHHDAQQ
Summary
Uniprot
D7F172
D7F163
D7F161
D7F168
A0A2W1BGD9
D7F162
+ More
D7F167 A0A3S2L0R7 D7F175 D5LB39 A0A402E714 A0A402ETA8 A0A402F4J0 A0A402EU50 A0A402EWZ2 A0A402F3F5 A0A402FRB1 A0A402E7T0 A0A402F8Q6 A0A402EYG3 A0A402EWW2 A0A402F4T1 A0A402E7S7 A0A402F6F8 A0A402ECQ6 A0A3Q1M4E7 A0A402EWK5 A0A402EPJ2 A0A402F693 A0A402EL59 A0A402F382 A0A402E7N2 A0A2G8JE23 A0A402F971 A0A402EQ09 A0A402ESY2 A0A402F7I8 A0A402ELI2 A0A402E871 A0A402EWK9 A0A402ET45 A0A402EV46 A0A402EH83 A0A3Q1MCX3 A0A402F1W2 A0A402EWV0 A0A3Q1LLH3 A0A402F9T4 A0A3Q1M7G0 A0A402E7Z5 A0A402EWI8 A0A402EBJ8 W5NMM5 A0A402EUK1 A0A402EU33 A0A2H6KKM9 A0A402E5M8 A0A402EL35 A0A402ELM7 A0A402EB12 A0A3Q1M4J9 A0A402EKB1 A0A402F3Q6 A0A402E825 A0A3Q1N3Y9 A0A402E768 A0A3Q1MJK8 A0A402F406 A0A402FC20 A0A3Q1MMS2 A0A3Q1LNR9 F6R5D0 A0A402EWI9 A0A3Q1MN71 A0A402E7R7 A0A402F9A7 A0A402ECS2 A0A402EBP1 A0A402FAG4 A0A3Q1NB85 A0A3Q1MDY8 A0A402EVG7 A0A3Q1MGA3 A0A2H1VU13 A0A3Q1MSR0 A0A3Q1MJR3 A0A402EBC2 A0A402EWU2 A0A402EWP9 A0A402ET28 A0A402EKM3 A0A3Q1LKN4 A0A402F575 A0A402EZW9 A0A3Q1MVF7 A0A402EBI2 A0A402EKC4 A0A402ET04 A0A3Q1MXT5 A0A402F125
D7F167 A0A3S2L0R7 D7F175 D5LB39 A0A402E714 A0A402ETA8 A0A402F4J0 A0A402EU50 A0A402EWZ2 A0A402F3F5 A0A402FRB1 A0A402E7T0 A0A402F8Q6 A0A402EYG3 A0A402EWW2 A0A402F4T1 A0A402E7S7 A0A402F6F8 A0A402ECQ6 A0A3Q1M4E7 A0A402EWK5 A0A402EPJ2 A0A402F693 A0A402EL59 A0A402F382 A0A402E7N2 A0A2G8JE23 A0A402F971 A0A402EQ09 A0A402ESY2 A0A402F7I8 A0A402ELI2 A0A402E871 A0A402EWK9 A0A402ET45 A0A402EV46 A0A402EH83 A0A3Q1MCX3 A0A402F1W2 A0A402EWV0 A0A3Q1LLH3 A0A402F9T4 A0A3Q1M7G0 A0A402E7Z5 A0A402EWI8 A0A402EBJ8 W5NMM5 A0A402EUK1 A0A402EU33 A0A2H6KKM9 A0A402E5M8 A0A402EL35 A0A402ELM7 A0A402EB12 A0A3Q1M4J9 A0A402EKB1 A0A402F3Q6 A0A402E825 A0A3Q1N3Y9 A0A402E768 A0A3Q1MJK8 A0A402F406 A0A402FC20 A0A3Q1MMS2 A0A3Q1LNR9 F6R5D0 A0A402EWI9 A0A3Q1MN71 A0A402E7R7 A0A402F9A7 A0A402ECS2 A0A402EBP1 A0A402FAG4 A0A3Q1NB85 A0A3Q1MDY8 A0A402EVG7 A0A3Q1MGA3 A0A2H1VU13 A0A3Q1MSR0 A0A3Q1MJR3 A0A402EBC2 A0A402EWU2 A0A402EWP9 A0A402ET28 A0A402EKM3 A0A3Q1LKN4 A0A402F575 A0A402EZW9 A0A3Q1MVF7 A0A402EBI2 A0A402EKC4 A0A402ET04 A0A3Q1MXT5 A0A402F125
EMBL
FJ265557
ADI61825.1
FJ265548
ADI61816.1
FJ265546
ADI61814.1
+ More
FJ265553 ADI61821.1 KZ150203 PZC72267.1 FJ265547 ADI61815.1 FJ265552 ADI61820.1 RSAL01000289 RVE42931.1 FJ265560 ADI61828.1 GU815090 ADF18553.1 BDOT01000002 GCF39995.1 BDOT01000039 GCF47371.1 BDOT01000104 GCF51438.1 BDOT01000043 GCF47717.1 BDOT01000015 BDOT01000055 BDOT01000148 GCF44364.1 GCF48654.1 GCF52988.1 BDOT01000089 GCF50820.1 BDOT01000474 GCF58974.1 GCF40240.1 BDOT01000143 GCF52806.1 BDOT01000062 GCF49174.1 BDOT01000054 GCF48637.1 BDOT01000105 GCF51451.1 GCF40256.1 BDOT01000116 GCF51880.1 BDOT01000006 GCF41961.1 GCF48602.1 BDOT01000028 GCF46132.1 BDOT01000112 GCF51757.1 BDOT01000020 GCF44935.1 BDOT01000090 GCF50913.1 GCF40190.1 MRZV01002322 PIK33998.1 BDOT01000149 GCF53002.1 BDOT01000029 GCF46280.1 GCF47356.1 BDOT01000130 GCF52369.1 BDOT01000022 GCF45060.1 GCF40380.1 GCF48608.1 GCF47377.1 BDOT01000048 GCF48010.1 BDOT01000011 GCF43541.1 BDOT01000083 GCF50492.1 BDOT01000056 GCF48705.1 BDOT01000156 GCF53186.1 GCF40308.1 GCF48559.1 BDOT01000005 GCF41553.1 AHAT01022938 BDOT01000045 GCF47820.1 GCF47708.1 BDSA01000081 GBE63537.1 BDOT01000001 GCF39495.1 GCF44897.1 GCF45061.1 GCF41419.1 BDOT01000017 GCF44637.1 BDOT01000094 GCF51029.1 GCF40355.1 GCF39993.1 GCF51020.1 BDOT01000183 GCF53956.1 GCF48586.1 GCF40259.1 BDOT01000150 GCF53030.1 GCF41910.1 GCF41584.1 BDOT01000165 GCF53431.1 BDOT01000049 GCF48208.1 ODYU01004401 SOQ44236.1 GCF41464.1 GCF48696.1 GCF48631.1 GCF47373.1 BDOT01000018 GCF44752.1 GCF51450.1 BDOT01000069 GCF49674.1 GCF41524.1 GCF44634.1 BDOT01000038 GCF47332.1 BDOT01000077 GCF50202.1
FJ265553 ADI61821.1 KZ150203 PZC72267.1 FJ265547 ADI61815.1 FJ265552 ADI61820.1 RSAL01000289 RVE42931.1 FJ265560 ADI61828.1 GU815090 ADF18553.1 BDOT01000002 GCF39995.1 BDOT01000039 GCF47371.1 BDOT01000104 GCF51438.1 BDOT01000043 GCF47717.1 BDOT01000015 BDOT01000055 BDOT01000148 GCF44364.1 GCF48654.1 GCF52988.1 BDOT01000089 GCF50820.1 BDOT01000474 GCF58974.1 GCF40240.1 BDOT01000143 GCF52806.1 BDOT01000062 GCF49174.1 BDOT01000054 GCF48637.1 BDOT01000105 GCF51451.1 GCF40256.1 BDOT01000116 GCF51880.1 BDOT01000006 GCF41961.1 GCF48602.1 BDOT01000028 GCF46132.1 BDOT01000112 GCF51757.1 BDOT01000020 GCF44935.1 BDOT01000090 GCF50913.1 GCF40190.1 MRZV01002322 PIK33998.1 BDOT01000149 GCF53002.1 BDOT01000029 GCF46280.1 GCF47356.1 BDOT01000130 GCF52369.1 BDOT01000022 GCF45060.1 GCF40380.1 GCF48608.1 GCF47377.1 BDOT01000048 GCF48010.1 BDOT01000011 GCF43541.1 BDOT01000083 GCF50492.1 BDOT01000056 GCF48705.1 BDOT01000156 GCF53186.1 GCF40308.1 GCF48559.1 BDOT01000005 GCF41553.1 AHAT01022938 BDOT01000045 GCF47820.1 GCF47708.1 BDSA01000081 GBE63537.1 BDOT01000001 GCF39495.1 GCF44897.1 GCF45061.1 GCF41419.1 BDOT01000017 GCF44637.1 BDOT01000094 GCF51029.1 GCF40355.1 GCF39993.1 GCF51020.1 BDOT01000183 GCF53956.1 GCF48586.1 GCF40259.1 BDOT01000150 GCF53030.1 GCF41910.1 GCF41584.1 BDOT01000165 GCF53431.1 BDOT01000049 GCF48208.1 ODYU01004401 SOQ44236.1 GCF41464.1 GCF48696.1 GCF48631.1 GCF47373.1 BDOT01000018 GCF44752.1 GCF51450.1 BDOT01000069 GCF49674.1 GCF41524.1 GCF44634.1 BDOT01000038 GCF47332.1 BDOT01000077 GCF50202.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
D7F172
D7F163
D7F161
D7F168
A0A2W1BGD9
D7F162
+ More
D7F167 A0A3S2L0R7 D7F175 D5LB39 A0A402E714 A0A402ETA8 A0A402F4J0 A0A402EU50 A0A402EWZ2 A0A402F3F5 A0A402FRB1 A0A402E7T0 A0A402F8Q6 A0A402EYG3 A0A402EWW2 A0A402F4T1 A0A402E7S7 A0A402F6F8 A0A402ECQ6 A0A3Q1M4E7 A0A402EWK5 A0A402EPJ2 A0A402F693 A0A402EL59 A0A402F382 A0A402E7N2 A0A2G8JE23 A0A402F971 A0A402EQ09 A0A402ESY2 A0A402F7I8 A0A402ELI2 A0A402E871 A0A402EWK9 A0A402ET45 A0A402EV46 A0A402EH83 A0A3Q1MCX3 A0A402F1W2 A0A402EWV0 A0A3Q1LLH3 A0A402F9T4 A0A3Q1M7G0 A0A402E7Z5 A0A402EWI8 A0A402EBJ8 W5NMM5 A0A402EUK1 A0A402EU33 A0A2H6KKM9 A0A402E5M8 A0A402EL35 A0A402ELM7 A0A402EB12 A0A3Q1M4J9 A0A402EKB1 A0A402F3Q6 A0A402E825 A0A3Q1N3Y9 A0A402E768 A0A3Q1MJK8 A0A402F406 A0A402FC20 A0A3Q1MMS2 A0A3Q1LNR9 F6R5D0 A0A402EWI9 A0A3Q1MN71 A0A402E7R7 A0A402F9A7 A0A402ECS2 A0A402EBP1 A0A402FAG4 A0A3Q1NB85 A0A3Q1MDY8 A0A402EVG7 A0A3Q1MGA3 A0A2H1VU13 A0A3Q1MSR0 A0A3Q1MJR3 A0A402EBC2 A0A402EWU2 A0A402EWP9 A0A402ET28 A0A402EKM3 A0A3Q1LKN4 A0A402F575 A0A402EZW9 A0A3Q1MVF7 A0A402EBI2 A0A402EKC4 A0A402ET04 A0A3Q1MXT5 A0A402F125
D7F167 A0A3S2L0R7 D7F175 D5LB39 A0A402E714 A0A402ETA8 A0A402F4J0 A0A402EU50 A0A402EWZ2 A0A402F3F5 A0A402FRB1 A0A402E7T0 A0A402F8Q6 A0A402EYG3 A0A402EWW2 A0A402F4T1 A0A402E7S7 A0A402F6F8 A0A402ECQ6 A0A3Q1M4E7 A0A402EWK5 A0A402EPJ2 A0A402F693 A0A402EL59 A0A402F382 A0A402E7N2 A0A2G8JE23 A0A402F971 A0A402EQ09 A0A402ESY2 A0A402F7I8 A0A402ELI2 A0A402E871 A0A402EWK9 A0A402ET45 A0A402EV46 A0A402EH83 A0A3Q1MCX3 A0A402F1W2 A0A402EWV0 A0A3Q1LLH3 A0A402F9T4 A0A3Q1M7G0 A0A402E7Z5 A0A402EWI8 A0A402EBJ8 W5NMM5 A0A402EUK1 A0A402EU33 A0A2H6KKM9 A0A402E5M8 A0A402EL35 A0A402ELM7 A0A402EB12 A0A3Q1M4J9 A0A402EKB1 A0A402F3Q6 A0A402E825 A0A3Q1N3Y9 A0A402E768 A0A3Q1MJK8 A0A402F406 A0A402FC20 A0A3Q1MMS2 A0A3Q1LNR9 F6R5D0 A0A402EWI9 A0A3Q1MN71 A0A402E7R7 A0A402F9A7 A0A402ECS2 A0A402EBP1 A0A402FAG4 A0A3Q1NB85 A0A3Q1MDY8 A0A402EVG7 A0A3Q1MGA3 A0A2H1VU13 A0A3Q1MSR0 A0A3Q1MJR3 A0A402EBC2 A0A402EWU2 A0A402EWP9 A0A402ET28 A0A402EKM3 A0A3Q1LKN4 A0A402F575 A0A402EZW9 A0A3Q1MVF7 A0A402EBI2 A0A402EKC4 A0A402ET04 A0A3Q1MXT5 A0A402F125
Ontologies
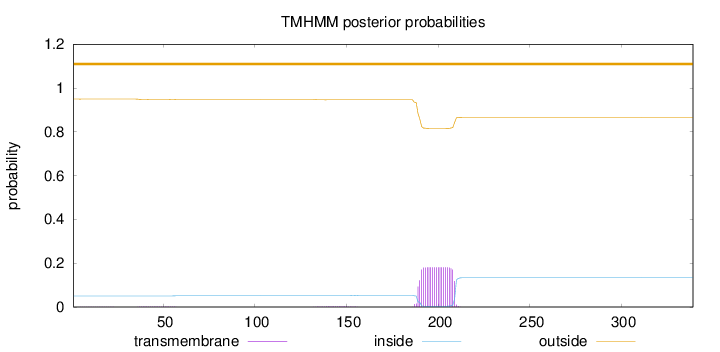
Topology
Length:
339
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.70768
Exp number, first 60 AAs:
0.04073
Total prob of N-in:
0.05035
outside
1 - 339
Population Genetic Test Statistics
Pi
25.176171
Theta
15.488208
Tajima's D
-1.259192
CLR
1166.554903
CSRT
0.0928953552322384
Interpretation
Uncertain
