Gene
KWMTBOMO08966
Pre Gene Modal
BGIBMGA007794
Annotation
PREDICTED:_potassium_voltage-gated_channel_subfamily_H_member_6-like_isoform_X3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.575
Sequence
CDS
ATGGAACGTAAAGGACCTTACACGGAGATGAACAATACATTAACTCAGGCAGTTCTGTTTAACTCATGTCGCAGTCTTTGCAACAGAGGACAGGTACTCTCACTGGGAGCGACGGGTGGGCATAAAGGCACAACGCAGCCATCTCATCCATGCACTATCCTCCACCATTCTCCTTATAAGGCCGCTTGGGATTGGTTGATACTAATATTGGTTATCTACACGGCAATATTTACTCCCTACGTAGCTGCCTTTCAACTAAATGAACCCGATTTCGACAAGCGTTCTCGAAGCTTCGGCGAAGATCCCATTGTAGTTATAGATATGATAGTCGACGTGACATTTATAATAGACATCTTAATAAACTTTCGAACAACATATGTGAACGCTGCTGACGAAGTGGAATCCGATCCTGCGAAAATTGCCATGCACTATCTTCGTGGCTGGTTCGTCATAGACCTCGTAGCTGCTATTCCCTTCGATCTTCTGCTGTTCGGCACCGACACCGATGAAACGACAACTCTCATCGGTTTACTGAAAACAGCGCGCTTGTTGCGACTAGTTAGAGTCGCTCGAAAAATTGACAGATACTCGGAATATGGCACCGCTGTCCTTCTATTGCTCATGGCTACGTTTGTTCTCATCGCACATTGGCTGGCGTGTTTATGGTATGCTATCGGGAGTTGGGAGCGACCGCAGCTACACGCTCCTATAGGTTGGTTAGATGCACTCGCTAACGATGTGCAAGCTTCCTATGACAATTCAACCGGCCCCTCTATGAGATCTCGTTATATCACCGCTTTATATTTCACGTGCACTTCCTTGACGAGCGTCGGTTTTGGAAATGTAGCTCCTAACACAGACATGGAAAAAGGTTTCACGATATTCGTAATGCTCGTTGGCTCACTTATGTACGCGAGTATTTTTGGAAATGTTTCTGCGATTATACAAAGACTGTATTCTGGAACAGCACGATACCATACACAAATACTTAGAGTACGAGAATTTATCAGATTTCATCAGATAACCAACCCCCTAAGACAACGCCTAGAAGAATATTTCCAACACGCGTGGAGCTATACAAATGGCATCGATACATCGAGCGTCCTGAAAGGATTCCCCGAGTGCCTCCAAGCTGATATCTGCCTCCACTTGAATCGTAATTTACTTGCCAGCTGTTCAGCCTTTGAAGGCGCCAGTCCTGGATGCTTAAGAGCCCTCTCCCTAAGATTCAAGACAACTCACGCTCCACCCGGTGAAACCCTAGTTCATAGAGGAGACGTTCTGACCTCCCTATACTTTATTTCGCGAGGATCCATTGAAATACTCAAAGATGACATAGTCATGGCCATTCTCGGGAAAGATGACATTTTCGGCGAGAACCCGTGCATTCATGGAACAGTCGGTCGTAGTAACTGCCGCGTGCGCGCTCTCACGTATTGCGATTTGCATAGAGTTCATAGAGACGACCTGCTAGACGTACTGGACTTCTATCCTGAATTCCGCAACTCCTTCGTTAATAACTTGGAGATAACTTACAACATGCGTGACGAGGAGTTAGGTGGTATAGAGCCGCGACGCCGGATGCGCTACGAGAATCCTTGCGACGCGCGTCTCCTGCGCGAAGCGTACGCGCGGACAACACGCCGTCCCCACACCGAGAACTTCGCTGAAAAAATGGTACGCGCTTCAGTTTTATGTAGAATTGCTTTCGAACGCCGATGGTCAGCGCTGCTCACGGACGCCATCACTTAA
Protein
MERKGPYTEMNNTLTQAVLFNSCRSLCNRGQVLSLGATGGHKGTTQPSHPCTILHHSPYKAAWDWLILILVIYTAIFTPYVAAFQLNEPDFDKRSRSFGEDPIVVIDMIVDVTFIIDILINFRTTYVNAADEVESDPAKIAMHYLRGWFVIDLVAAIPFDLLLFGTDTDETTTLIGLLKTARLLRLVRVARKIDRYSEYGTAVLLLLMATFVLIAHWLACLWYAIGSWERPQLHAPIGWLDALANDVQASYDNSTGPSMRSRYITALYFTCTSLTSVGFGNVAPNTDMEKGFTIFVMLVGSLMYASIFGNVSAIIQRLYSGTARYHTQILRVREFIRFHQITNPLRQRLEEYFQHAWSYTNGIDTSSVLKGFPECLQADICLHLNRNLLASCSAFEGASPGCLRALSLRFKTTHAPPGETLVHRGDVLTSLYFISRGSIEILKDDIVMAILGKDDIFGENPCIHGTVGRSNCRVRALTYCDLHRVHRDDLLDVLDFYPEFRNSFVNNLEITYNMRDEELGGIEPRRRMRYENPCDARLLREAYARTTRRPHTENFAEKMVRASVLCRIAFERRWSALLTDAIT
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
A0A0K0YB78
A0A2H1VXM3
A0A2A4J7U4
A0A2A4J688
A0A212FDK2
A0A3S2NGE0
+ More
A0A0K0YB79 A0A194PIS4 H9JE47 A0A194RCD5 A0A0T6BGA5 A0A1Y1NE51 A0A1Y1NDV1 A0A1L8DEW9 A0A151XGX8 A0A151IMH8 A0A2U8JGI1 A0A195DUQ9 D2A5B0 A0A195ETC9 A0A067QGI0 A0A195AYI9 N6UAH6 A0A0C9QN59 A0A0C9RBX3 A0A0C9R3J2 A0A158NSX7 A0A182JTW2 E2ADF1 A0A146KJC2 A0A0A9WVE9 A0A182RB17 A0A023PMM1 A0A182WIY8 A0A0P6DWS5 A0A0P5ZUZ6 A0A0P5ZWM0 A0A0P4XWC1 A0A182V624 A0A0P6E2X5 A0A0P5L4F9 A0A0P6EC30 A0A026WZI3 A0A453Z1J7 A0A0J7NX39 A0A0P5TSS9 A0A0P5ELC4 A0A0P5UDZ1 A0A0N8A1M2 A0A0N8B3L6 A0A182IX06 Q7QJX3 A0A0P6FHR2 A0A0P5AVD2 A0A0P4YM11 A0A0P6FQF6 A0A0P5DNP5 A0A0P5TG72 A0A1J1IU73 A0A0P5ET03 A0A0P6EQ94 A0A0P6DV70 A0A0P5T6C2 A0A0P4YH50 A0A0P5ZTE1 A0A1B0FJW3 A0A0P4ZBC2 A0A0P5NDU8 A0A0P6AIH7 A0A0P6IN14 W5JKR4 A0A0N8EQK6 A0A453Z1J4 A0A0P5VVN6 A0A1D2NMC7 A0A0P5WA93 A0A0P5AEJ3 A0A0P4Y2W7 A0A0N8CXK9 A0A453YZR4 A0A0P6F9G9 A0A0P6HAE1 A0A1B0A992 A0A165A9H5 A0A0P5JYP0 A0A0P5MK54 A0A1A9Y6J2 A0A1A9UJ19 A0A0L0C9D9 A0A1B0BIA8 A0A0P6GSP9 A0A0P5MMS7 A0A182I3W2 A0A1A9WP63 A0A453YZW9 A0A154P9R5 A0A2H8TZV5 A0A182M7I3 A0A453YZZ0 A0A3L8DS60
A0A0K0YB79 A0A194PIS4 H9JE47 A0A194RCD5 A0A0T6BGA5 A0A1Y1NE51 A0A1Y1NDV1 A0A1L8DEW9 A0A151XGX8 A0A151IMH8 A0A2U8JGI1 A0A195DUQ9 D2A5B0 A0A195ETC9 A0A067QGI0 A0A195AYI9 N6UAH6 A0A0C9QN59 A0A0C9RBX3 A0A0C9R3J2 A0A158NSX7 A0A182JTW2 E2ADF1 A0A146KJC2 A0A0A9WVE9 A0A182RB17 A0A023PMM1 A0A182WIY8 A0A0P6DWS5 A0A0P5ZUZ6 A0A0P5ZWM0 A0A0P4XWC1 A0A182V624 A0A0P6E2X5 A0A0P5L4F9 A0A0P6EC30 A0A026WZI3 A0A453Z1J7 A0A0J7NX39 A0A0P5TSS9 A0A0P5ELC4 A0A0P5UDZ1 A0A0N8A1M2 A0A0N8B3L6 A0A182IX06 Q7QJX3 A0A0P6FHR2 A0A0P5AVD2 A0A0P4YM11 A0A0P6FQF6 A0A0P5DNP5 A0A0P5TG72 A0A1J1IU73 A0A0P5ET03 A0A0P6EQ94 A0A0P6DV70 A0A0P5T6C2 A0A0P4YH50 A0A0P5ZTE1 A0A1B0FJW3 A0A0P4ZBC2 A0A0P5NDU8 A0A0P6AIH7 A0A0P6IN14 W5JKR4 A0A0N8EQK6 A0A453Z1J4 A0A0P5VVN6 A0A1D2NMC7 A0A0P5WA93 A0A0P5AEJ3 A0A0P4Y2W7 A0A0N8CXK9 A0A453YZR4 A0A0P6F9G9 A0A0P6HAE1 A0A1B0A992 A0A165A9H5 A0A0P5JYP0 A0A0P5MK54 A0A1A9Y6J2 A0A1A9UJ19 A0A0L0C9D9 A0A1B0BIA8 A0A0P6GSP9 A0A0P5MMS7 A0A182I3W2 A0A1A9WP63 A0A453YZW9 A0A154P9R5 A0A2H8TZV5 A0A182M7I3 A0A453YZZ0 A0A3L8DS60
Pubmed
EMBL
KR081241
AKS48231.1
ODYU01005069
SOQ45585.1
NWSH01002828
PCG67472.1
+ More
PCG67471.1 AGBW02009058 OWR51778.1 RSAL01000049 RVE50390.1 KR081243 AKS48233.1 KQ459602 KPI93321.1 BABH01002584 BABH01002585 KQ460615 KPJ13566.1 LJIG01000670 KRT86270.1 GEZM01005648 JAV96009.1 GEZM01005647 JAV96011.1 GFDF01009149 JAV04935.1 KQ982138 KYQ59656.1 KQ977047 KYN06092.1 MF360102 AWK67800.1 KQ980390 KYN16294.1 KQ971345 EFA05339.2 KQ981989 KYN31167.1 KK853474 KDR07360.1 KQ976711 KYM77029.1 APGK01036682 APGK01036683 APGK01036684 APGK01036685 APGK01036686 KB740941 ENN77636.1 GBYB01002087 JAG71854.1 GBYB01010627 JAG80394.1 GBYB01001381 JAG71148.1 ADTU01025304 ADTU01025305 ADTU01025306 ADTU01025307 ADTU01025308 ADTU01025309 ADTU01025310 ADTU01025311 ADTU01025312 ADTU01025313 GL438749 EFN68567.1 GDHC01021948 JAP96680.1 GBHO01032198 JAG11406.1 KJ493818 AHX25744.1 GDIQ01072928 JAN21809.1 GDIP01039434 JAM64281.1 GDIP01038333 JAM65382.1 GDIP01237068 JAI86333.1 GDIQ01083377 JAN11360.1 GDIQ01175342 JAK76383.1 GDIQ01066235 JAN28502.1 KK107054 EZA61480.1 LBMM01001072 KMQ96980.1 GDIP01125017 JAL78697.1 GDIP01146515 JAJ76887.1 GDIP01114419 GDIP01114418 JAL89295.1 GDIP01191361 JAJ32041.1 GDIQ01216131 JAK35594.1 AAAB01008807 EAA04103.4 GDIQ01057133 JAN37604.1 GDIP01193611 JAJ29791.1 GDIP01235461 JAI87940.1 GDIQ01044742 JAN49995.1 GDIP01153819 JAJ69583.1 GDIP01130374 JAL73340.1 CVRI01000059 CRL03250.1 GDIP01155571 JAJ67831.1 GDIQ01072927 JAN21810.1 GDIQ01075729 JAN19008.1 GDIP01130373 JAL73341.1 GDIP01237070 JAI86331.1 GDIP01039435 JAM64280.1 CCAG010018850 GDIP01228110 JAI95291.1 GDIQ01143956 JAL07770.1 GDIP01028892 JAM74823.1 GDIQ01029150 JAN65587.1 ADMH02001258 ETN63364.1 GDIQ01003620 JAN91117.1 GDIP01094622 JAM09093.1 LJIJ01000008 ODN06245.1 GDIP01088973 JAM14742.1 GDIP01200866 JAJ22536.1 GDIP01237069 JAI86332.1 GDIP01088975 GDIP01088974 JAM14740.1 GDIQ01052691 JAN42046.1 GDIQ01034294 JAN60443.1 LRGB01000642 KZS17336.1 GDIQ01197132 JAK54593.1 GDIQ01156897 JAK94828.1 JRES01000737 KNC28850.1 JXJN01014865 JXJN01014866 JXJN01014867 GDIQ01030551 JAN64186.1 GDIQ01154405 JAK97320.1 APCN01000225 KQ434853 KZC08649.1 GFXV01008001 MBW19806.1 AXCM01001458 QOIP01000004 RLU23241.1
PCG67471.1 AGBW02009058 OWR51778.1 RSAL01000049 RVE50390.1 KR081243 AKS48233.1 KQ459602 KPI93321.1 BABH01002584 BABH01002585 KQ460615 KPJ13566.1 LJIG01000670 KRT86270.1 GEZM01005648 JAV96009.1 GEZM01005647 JAV96011.1 GFDF01009149 JAV04935.1 KQ982138 KYQ59656.1 KQ977047 KYN06092.1 MF360102 AWK67800.1 KQ980390 KYN16294.1 KQ971345 EFA05339.2 KQ981989 KYN31167.1 KK853474 KDR07360.1 KQ976711 KYM77029.1 APGK01036682 APGK01036683 APGK01036684 APGK01036685 APGK01036686 KB740941 ENN77636.1 GBYB01002087 JAG71854.1 GBYB01010627 JAG80394.1 GBYB01001381 JAG71148.1 ADTU01025304 ADTU01025305 ADTU01025306 ADTU01025307 ADTU01025308 ADTU01025309 ADTU01025310 ADTU01025311 ADTU01025312 ADTU01025313 GL438749 EFN68567.1 GDHC01021948 JAP96680.1 GBHO01032198 JAG11406.1 KJ493818 AHX25744.1 GDIQ01072928 JAN21809.1 GDIP01039434 JAM64281.1 GDIP01038333 JAM65382.1 GDIP01237068 JAI86333.1 GDIQ01083377 JAN11360.1 GDIQ01175342 JAK76383.1 GDIQ01066235 JAN28502.1 KK107054 EZA61480.1 LBMM01001072 KMQ96980.1 GDIP01125017 JAL78697.1 GDIP01146515 JAJ76887.1 GDIP01114419 GDIP01114418 JAL89295.1 GDIP01191361 JAJ32041.1 GDIQ01216131 JAK35594.1 AAAB01008807 EAA04103.4 GDIQ01057133 JAN37604.1 GDIP01193611 JAJ29791.1 GDIP01235461 JAI87940.1 GDIQ01044742 JAN49995.1 GDIP01153819 JAJ69583.1 GDIP01130374 JAL73340.1 CVRI01000059 CRL03250.1 GDIP01155571 JAJ67831.1 GDIQ01072927 JAN21810.1 GDIQ01075729 JAN19008.1 GDIP01130373 JAL73341.1 GDIP01237070 JAI86331.1 GDIP01039435 JAM64280.1 CCAG010018850 GDIP01228110 JAI95291.1 GDIQ01143956 JAL07770.1 GDIP01028892 JAM74823.1 GDIQ01029150 JAN65587.1 ADMH02001258 ETN63364.1 GDIQ01003620 JAN91117.1 GDIP01094622 JAM09093.1 LJIJ01000008 ODN06245.1 GDIP01088973 JAM14742.1 GDIP01200866 JAJ22536.1 GDIP01237069 JAI86332.1 GDIP01088975 GDIP01088974 JAM14740.1 GDIQ01052691 JAN42046.1 GDIQ01034294 JAN60443.1 LRGB01000642 KZS17336.1 GDIQ01197132 JAK54593.1 GDIQ01156897 JAK94828.1 JRES01000737 KNC28850.1 JXJN01014865 JXJN01014866 JXJN01014867 GDIQ01030551 JAN64186.1 GDIQ01154405 JAK97320.1 APCN01000225 KQ434853 KZC08649.1 GFXV01008001 MBW19806.1 AXCM01001458 QOIP01000004 RLU23241.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053268
UP000005204
UP000053240
+ More
UP000075809 UP000078542 UP000078492 UP000007266 UP000078541 UP000027135 UP000078540 UP000019118 UP000005205 UP000075881 UP000000311 UP000075900 UP000075920 UP000075903 UP000053097 UP000036403 UP000075880 UP000007062 UP000183832 UP000092444 UP000000673 UP000094527 UP000092445 UP000076858 UP000092443 UP000078200 UP000037069 UP000092460 UP000075840 UP000091820 UP000076502 UP000075883 UP000279307
UP000075809 UP000078542 UP000078492 UP000007266 UP000078541 UP000027135 UP000078540 UP000019118 UP000005205 UP000075881 UP000000311 UP000075900 UP000075920 UP000075903 UP000053097 UP000036403 UP000075880 UP000007062 UP000183832 UP000092444 UP000000673 UP000094527 UP000092445 UP000076858 UP000092443 UP000078200 UP000037069 UP000092460 UP000075840 UP000091820 UP000076502 UP000075883 UP000279307
PRIDE
Pfam
Interpro
IPR018490
cNMP-bd-like
+ More
IPR014710 RmlC-like_jellyroll
IPR003967 K_chnl_volt-dep_ERG
IPR030172 KCNH6
IPR000595 cNMP-bd_dom
IPR003938 K_chnl_volt-dep_EAG/ELK/ERG
IPR027359 Volt_channel_dom_sf
IPR005821 Ion_trans_dom
IPR000014 PAS
IPR035965 PAS-like_dom_sf
IPR006202 Neur_chan_lig-bd
IPR038105 Kif23_Arf-bd_sf
IPR032384 Kif23_Arf-bd
IPR019821 Kinesin_motor_CS
IPR018000 Neurotransmitter_ion_chnl_CS
IPR036961 Kinesin_motor_dom_sf
IPR036734 Neur_chan_lig-bd_sf
IPR036719 Neuro-gated_channel_TM_sf
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR002130 Cyclophilin-type_PPIase_dom
IPR029000 Cyclophilin-like_dom_sf
IPR006029 Neurotrans-gated_channel_TM
IPR020892 Cyclophilin-type_PPIase_CS
IPR013122 PKD1_2_channel
IPR036013 Band_7/SPFH_dom_sf
IPR032435 Band_7_C
IPR001107 Band_7
IPR014710 RmlC-like_jellyroll
IPR003967 K_chnl_volt-dep_ERG
IPR030172 KCNH6
IPR000595 cNMP-bd_dom
IPR003938 K_chnl_volt-dep_EAG/ELK/ERG
IPR027359 Volt_channel_dom_sf
IPR005821 Ion_trans_dom
IPR000014 PAS
IPR035965 PAS-like_dom_sf
IPR006202 Neur_chan_lig-bd
IPR038105 Kif23_Arf-bd_sf
IPR032384 Kif23_Arf-bd
IPR019821 Kinesin_motor_CS
IPR018000 Neurotransmitter_ion_chnl_CS
IPR036961 Kinesin_motor_dom_sf
IPR036734 Neur_chan_lig-bd_sf
IPR036719 Neuro-gated_channel_TM_sf
IPR027417 P-loop_NTPase
IPR001752 Kinesin_motor_dom
IPR002130 Cyclophilin-type_PPIase_dom
IPR029000 Cyclophilin-like_dom_sf
IPR006029 Neurotrans-gated_channel_TM
IPR020892 Cyclophilin-type_PPIase_CS
IPR013122 PKD1_2_channel
IPR036013 Band_7/SPFH_dom_sf
IPR032435 Band_7_C
IPR001107 Band_7
SUPFAM
ProteinModelPortal
A0A0K0YB78
A0A2H1VXM3
A0A2A4J7U4
A0A2A4J688
A0A212FDK2
A0A3S2NGE0
+ More
A0A0K0YB79 A0A194PIS4 H9JE47 A0A194RCD5 A0A0T6BGA5 A0A1Y1NE51 A0A1Y1NDV1 A0A1L8DEW9 A0A151XGX8 A0A151IMH8 A0A2U8JGI1 A0A195DUQ9 D2A5B0 A0A195ETC9 A0A067QGI0 A0A195AYI9 N6UAH6 A0A0C9QN59 A0A0C9RBX3 A0A0C9R3J2 A0A158NSX7 A0A182JTW2 E2ADF1 A0A146KJC2 A0A0A9WVE9 A0A182RB17 A0A023PMM1 A0A182WIY8 A0A0P6DWS5 A0A0P5ZUZ6 A0A0P5ZWM0 A0A0P4XWC1 A0A182V624 A0A0P6E2X5 A0A0P5L4F9 A0A0P6EC30 A0A026WZI3 A0A453Z1J7 A0A0J7NX39 A0A0P5TSS9 A0A0P5ELC4 A0A0P5UDZ1 A0A0N8A1M2 A0A0N8B3L6 A0A182IX06 Q7QJX3 A0A0P6FHR2 A0A0P5AVD2 A0A0P4YM11 A0A0P6FQF6 A0A0P5DNP5 A0A0P5TG72 A0A1J1IU73 A0A0P5ET03 A0A0P6EQ94 A0A0P6DV70 A0A0P5T6C2 A0A0P4YH50 A0A0P5ZTE1 A0A1B0FJW3 A0A0P4ZBC2 A0A0P5NDU8 A0A0P6AIH7 A0A0P6IN14 W5JKR4 A0A0N8EQK6 A0A453Z1J4 A0A0P5VVN6 A0A1D2NMC7 A0A0P5WA93 A0A0P5AEJ3 A0A0P4Y2W7 A0A0N8CXK9 A0A453YZR4 A0A0P6F9G9 A0A0P6HAE1 A0A1B0A992 A0A165A9H5 A0A0P5JYP0 A0A0P5MK54 A0A1A9Y6J2 A0A1A9UJ19 A0A0L0C9D9 A0A1B0BIA8 A0A0P6GSP9 A0A0P5MMS7 A0A182I3W2 A0A1A9WP63 A0A453YZW9 A0A154P9R5 A0A2H8TZV5 A0A182M7I3 A0A453YZZ0 A0A3L8DS60
A0A0K0YB79 A0A194PIS4 H9JE47 A0A194RCD5 A0A0T6BGA5 A0A1Y1NE51 A0A1Y1NDV1 A0A1L8DEW9 A0A151XGX8 A0A151IMH8 A0A2U8JGI1 A0A195DUQ9 D2A5B0 A0A195ETC9 A0A067QGI0 A0A195AYI9 N6UAH6 A0A0C9QN59 A0A0C9RBX3 A0A0C9R3J2 A0A158NSX7 A0A182JTW2 E2ADF1 A0A146KJC2 A0A0A9WVE9 A0A182RB17 A0A023PMM1 A0A182WIY8 A0A0P6DWS5 A0A0P5ZUZ6 A0A0P5ZWM0 A0A0P4XWC1 A0A182V624 A0A0P6E2X5 A0A0P5L4F9 A0A0P6EC30 A0A026WZI3 A0A453Z1J7 A0A0J7NX39 A0A0P5TSS9 A0A0P5ELC4 A0A0P5UDZ1 A0A0N8A1M2 A0A0N8B3L6 A0A182IX06 Q7QJX3 A0A0P6FHR2 A0A0P5AVD2 A0A0P4YM11 A0A0P6FQF6 A0A0P5DNP5 A0A0P5TG72 A0A1J1IU73 A0A0P5ET03 A0A0P6EQ94 A0A0P6DV70 A0A0P5T6C2 A0A0P4YH50 A0A0P5ZTE1 A0A1B0FJW3 A0A0P4ZBC2 A0A0P5NDU8 A0A0P6AIH7 A0A0P6IN14 W5JKR4 A0A0N8EQK6 A0A453Z1J4 A0A0P5VVN6 A0A1D2NMC7 A0A0P5WA93 A0A0P5AEJ3 A0A0P4Y2W7 A0A0N8CXK9 A0A453YZR4 A0A0P6F9G9 A0A0P6HAE1 A0A1B0A992 A0A165A9H5 A0A0P5JYP0 A0A0P5MK54 A0A1A9Y6J2 A0A1A9UJ19 A0A0L0C9D9 A0A1B0BIA8 A0A0P6GSP9 A0A0P5MMS7 A0A182I3W2 A0A1A9WP63 A0A453YZW9 A0A154P9R5 A0A2H8TZV5 A0A182M7I3 A0A453YZZ0 A0A3L8DS60
PDB
5VA2
E-value=3.69039e-172,
Score=1554
Ontologies
GO
PANTHER
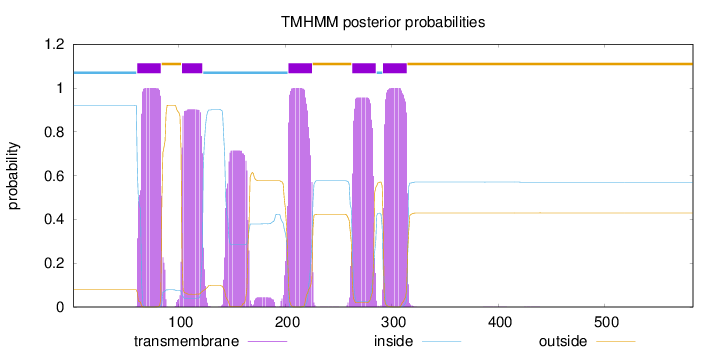
Topology
Length:
583
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
121.6613
Exp number, first 60 AAs:
0.00534
Total prob of N-in:
0.92164
inside
1 - 60
TMhelix
61 - 83
outside
84 - 102
TMhelix
103 - 122
inside
123 - 202
TMhelix
203 - 225
outside
226 - 262
TMhelix
263 - 285
inside
286 - 291
TMhelix
292 - 314
outside
315 - 583
Population Genetic Test Statistics
Pi
267.914171
Theta
191.072855
Tajima's D
1.219127
CLR
0.001532
CSRT
0.72186390680466
Interpretation
Uncertain
