Gene
KWMTBOMO08964 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007734
Annotation
PREDICTED:_bystin_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.173
Sequence
CDS
ATGGGTAAAGTTAAAAAGTTGAAGCCCTCACAAGTGGGAAAAAATTTAGCTCTTGCTGATCAAATTGAATCTGGAAATTCAGTCAAAATTAAAAATAGGACCAAAGATAGAAATAGGCACGATGAAGACGATGAGTTTGTAAAAGCAGATTTGTCTAAAAAAATTTTAAAAACTGCTAGAAGGCAACAAGCTGAACTAGAAGACAATGAGATTGGTCCGTCTCCAGCAAAACATGTCACTCTTGTGTCAAGCTTTAAAGATAAAGGATCAGATAGTTCAGATTCAGAGAAGGATGACTTGGAACCTGACACATACTATGATAACATTGAAATCAACGAAGCAGATGAAGAAGCACTCAAATTGTTCAAATCATCTAAAACAGAAAGAGTTAGAACTCTAGCTGATATTATAAAAGAAAAAATTACTGACAAACACACAGAACTACAGACTCAGTTCTCAGATGCAGAAACTTTAAAATTGCAGAATATTGATCCAAGAATAAAAACCATGTATGAGGGCGTAAGGGATGTACTCCAGAAATATAGATCAGGCAAATTACCGAAAGCTTTCAAAATGGTGCCACATTTGCAGAATTGGGAGCAAATTTTATACTTAACAGAGCCTACTACTTGGTCAGCAGCTGCTATGTATCAGGCAACTAGAATTTTTGCATCAAATCTTAAGGAGAAGATGGCTCAGAGGTTTTATAATTTAGTTTTACTACCACGTGTCAGAGATGACCTTGCTGAATACAAAAGGTTGAATTTCCACTTGTACCAAGCATTGAGGAAGGCCTTGTTCAAACCTGGTGCTTTTATGAAGGGAATTTTACTACCGTTATGTGAGGCTGGGGATTGTACGCTAAGAGAAGCTATAATTGTTGGATCTGTGCTGGCACGTAATTCAGTACCCGTTTTACATTCTAGTGCTGCTTTGTTAAAAATAGCTGAAATGGATTATAACGGTGCTAATTCAATTTTCCTGAGAATCCTTTTTGATAAAAAATATGCTCTTCCATATAGAGTTGTGGATGCTGTTGTTTTTCATTTTTTGAGGTTTCAATCTGAGGCTCGTGTATTACCTGTATTATGGCATCAAGCTCTATTAACGTTTGTACAGCGTTATAAGGCAGATATATCAACTGAGCAACGAGATGCACTTCTAGAGTTATTGAGAAAACAATATCATCCTACAATAACACAAGAGATTCGACGGGAGTTACAACAAGCTCAATGCAGAGATATTGAAGTAAATGAAGAAGCAACTAACATGATGTCAGTTGAATAA
Protein
MGKVKKLKPSQVGKNLALADQIESGNSVKIKNRTKDRNRHDEDDEFVKADLSKKILKTARRQQAELEDNEIGPSPAKHVTLVSSFKDKGSDSSDSEKDDLEPDTYYDNIEINEADEEALKLFKSSKTERVRTLADIIKEKITDKHTELQTQFSDAETLKLQNIDPRIKTMYEGVRDVLQKYRSGKLPKAFKMVPHLQNWEQILYLTEPTTWSAAAMYQATRIFASNLKEKMAQRFYNLVLLPRVRDDLAEYKRLNFHLYQALRKALFKPGAFMKGILLPLCEAGDCTLREAIIVGSVLARNSVPVLHSSAALLKIAEMDYNGANSIFLRILFDKKYALPYRVVDAVVFHFLRFQSEARVLPVLWHQALLTFVQRYKADISTEQRDALLELLRKQYHPTITQEIRRELQQAQCRDIEVNEEATNMMSVE
Summary
Uniprot
B9VTR7
H9JDY7
A0A2A4JSW8
A0A2H1VZC7
A0A194R8J0
A0A1E1WJA4
+ More
A0A194PQB0 S4PCD0 A0A212FDH3 A0A437BIY6 A0A2J7QLA3 A0A069DYY4 A0A067R4M0 A0A151X427 A0A224XQS3 A0A0P4VTY8 R4G3T9 A0A0V0G7S8 A0A195FSR5 A0A158NKZ3 A0A0N0U5Y7 F4WWT7 A0A023F8K2 A0A151JAB3 A0A0L7R3H7 A0A0J7L4N6 A0A154PDU5 D6WMH7 E2B315 E2AL69 A0A1Y1KQX7 A0A1B6EUR0 A0A232EJ63 K7IXM5 A0A087ZR91 A0A2A3E8T3 A0A1B6J988 A0A0C9PJ85 A0A151IMC5 A0A026WML5 A0A310SEA2 E0VFT7 A0A0A9Z1G7 A0A210PQY0 A0A2S2P4F3 A0A2S2PZU0 J9JM85 A0A2H8U2M6 C4WV65 A0A224YYH0 A0A131YKG8 L7M348 A0A0P4YMB0 A0A131XIW8 A0A293LEZ3 A0A1W4W8Y5 A0A023GKF5 G3MFJ5 A0A131XZI2 A0A1B6CJX0 V5I370 A0A0T6AZQ0 L7MIE1 K1Q864 B7Q8R6 N6TMN1 A0A182XV21 J3JY75 A0A182LUN6 A0A0N7ZPI7 A0A1L8E3L3 A0A0P5YEH7 A0A3Q1HNM2 A0A0P5DFC2 A0A0K8TT50 A0A1B0GK48 A0A182RVA5 Q0C738 A0A151NBS7 A0A0P5V7L6 A0A336MBH4 A0A182VRQ3 A0A0P4YL19 A0A182H887 A0A182JQM6 A0A1U7RWJ7 A0A182PQ05 A0A182TI11 A0A1S4EUN3 Q7QD29 A0A1S3K8C8 A0A3B3ZJC4 A0A2Y9D307 A0A182HTY1 Q7T3F8 A0A0P5TR74 R7U6A6 A0A023ES33 A8D881
A0A194PQB0 S4PCD0 A0A212FDH3 A0A437BIY6 A0A2J7QLA3 A0A069DYY4 A0A067R4M0 A0A151X427 A0A224XQS3 A0A0P4VTY8 R4G3T9 A0A0V0G7S8 A0A195FSR5 A0A158NKZ3 A0A0N0U5Y7 F4WWT7 A0A023F8K2 A0A151JAB3 A0A0L7R3H7 A0A0J7L4N6 A0A154PDU5 D6WMH7 E2B315 E2AL69 A0A1Y1KQX7 A0A1B6EUR0 A0A232EJ63 K7IXM5 A0A087ZR91 A0A2A3E8T3 A0A1B6J988 A0A0C9PJ85 A0A151IMC5 A0A026WML5 A0A310SEA2 E0VFT7 A0A0A9Z1G7 A0A210PQY0 A0A2S2P4F3 A0A2S2PZU0 J9JM85 A0A2H8U2M6 C4WV65 A0A224YYH0 A0A131YKG8 L7M348 A0A0P4YMB0 A0A131XIW8 A0A293LEZ3 A0A1W4W8Y5 A0A023GKF5 G3MFJ5 A0A131XZI2 A0A1B6CJX0 V5I370 A0A0T6AZQ0 L7MIE1 K1Q864 B7Q8R6 N6TMN1 A0A182XV21 J3JY75 A0A182LUN6 A0A0N7ZPI7 A0A1L8E3L3 A0A0P5YEH7 A0A3Q1HNM2 A0A0P5DFC2 A0A0K8TT50 A0A1B0GK48 A0A182RVA5 Q0C738 A0A151NBS7 A0A0P5V7L6 A0A336MBH4 A0A182VRQ3 A0A0P4YL19 A0A182H887 A0A182JQM6 A0A1U7RWJ7 A0A182PQ05 A0A182TI11 A0A1S4EUN3 Q7QD29 A0A1S3K8C8 A0A3B3ZJC4 A0A2Y9D307 A0A182HTY1 Q7T3F8 A0A0P5TR74 R7U6A6 A0A023ES33 A8D881
Pubmed
19121390
26354079
23622113
22118469
26334808
24845553
+ More
27129103 21347285 21719571 25474469 18362917 19820115 20798317 28004739 28648823 20075255 24508170 30249741 20566863 25401762 26823975 28812685 28797301 26830274 25576852 28049606 22216098 25765539 22992520 23537049 25244985 22516182 26369729 17510324 22293439 26483478 12364791 14747013 17210077 25463417 23254933 24945155
27129103 21347285 21719571 25474469 18362917 19820115 20798317 28004739 28648823 20075255 24508170 30249741 20566863 25401762 26823975 28812685 28797301 26830274 25576852 28049606 22216098 25765539 22992520 23537049 25244985 22516182 26369729 17510324 22293439 26483478 12364791 14747013 17210077 25463417 23254933 24945155
EMBL
FJ602777
ACM24343.1
BABH01002589
NWSH01000740
PCG74482.1
ODYU01005069
+ More
SOQ45584.1 KQ460615 KPJ13565.1 GDQN01003980 JAT87074.1 KQ459602 KPI93320.1 GAIX01005492 JAA87068.1 AGBW02009058 OWR51780.1 RSAL01000049 RVE50391.1 NEVH01013250 PNF29360.1 GBGD01001585 JAC87304.1 KK852751 KDR17157.1 KQ982557 KYQ55024.1 GFTR01006075 JAW10351.1 GDKW01001122 JAI55473.1 GAHY01001376 JAA76134.1 GECL01002356 JAP03768.1 KQ981281 KYN43342.1 ADTU01019058 KQ435747 KOX76494.1 GL888413 EGI61325.1 GBBI01001032 JAC17680.1 KQ979308 KYN21974.1 KQ414663 KOC65440.1 LBMM01000796 KMQ97469.1 KQ434885 KZC10066.1 KQ971343 EFA03295.1 GL445250 EFN89984.1 GL440492 EFN65824.1 GEZM01078253 GEZM01078252 JAV62821.1 GECZ01028067 JAS41702.1 NNAY01004075 OXU18407.1 AAZX01014366 KZ288322 PBC28120.1 GECU01011935 JAS95771.1 GBYB01001023 JAG70790.1 KQ977085 KYN05916.1 KK107152 QOIP01000007 EZA57153.1 RLU20694.1 KQ762520 OAD55765.1 DS235124 EEB12243.1 GBHO01006416 GBRD01013866 GDHC01014428 JAG37188.1 JAG51960.1 JAQ04201.1 NEDP02005553 OWF38862.1 GGMR01011681 MBY24300.1 GGMS01001815 MBY71018.1 ABLF02033113 GFXV01007993 MBW19798.1 AK341378 BAH71785.1 GFPF01007664 MAA18810.1 GEDV01008764 JAP79793.1 GACK01006742 JAA58292.1 GDIP01225243 GDIP01225242 JAI98158.1 GEFH01003090 JAP65491.1 GFWV01002582 MAA27312.1 GBBM01001052 JAC34366.1 JO840646 AEO32263.1 GEFM01003153 JAP72643.1 GEDC01023657 JAS13641.1 GANP01000321 JAB84147.1 LJIG01022450 KRT80530.1 GACK01001058 JAA63976.1 JH817849 EKC30098.1 ABJB010081940 ABJB010688253 DS885859 EEC15238.1 APGK01029293 KB740731 KB632226 ENN79288.1 ERL90308.1 BT128199 AEE63160.1 AXCM01000997 GDIP01225241 JAI98160.1 GFDF01000777 JAV13307.1 GDIP01058857 JAM44858.1 GDIP01158330 LRGB01002167 JAJ65072.1 KZS08857.1 GDAI01000059 JAI17544.1 AJWK01022559 CH477186 EAT48991.1 AKHW03003562 KYO34237.1 GDIP01104139 JAL99575.1 UFQS01000611 UFQT01000611 SSX05366.1 SSX25727.1 GDIP01229535 JAI93866.1 JXUM01117830 KQ566123 KXJ70378.1 AAAB01008859 EAA07612.5 APCN01000591 BC053134 AAH53134.1 GDIP01122792 JAL80922.1 AMQN01010124 KB307090 ELT99216.1 GAPW01001555 JAC12043.1 EU142260 ABV60396.1
SOQ45584.1 KQ460615 KPJ13565.1 GDQN01003980 JAT87074.1 KQ459602 KPI93320.1 GAIX01005492 JAA87068.1 AGBW02009058 OWR51780.1 RSAL01000049 RVE50391.1 NEVH01013250 PNF29360.1 GBGD01001585 JAC87304.1 KK852751 KDR17157.1 KQ982557 KYQ55024.1 GFTR01006075 JAW10351.1 GDKW01001122 JAI55473.1 GAHY01001376 JAA76134.1 GECL01002356 JAP03768.1 KQ981281 KYN43342.1 ADTU01019058 KQ435747 KOX76494.1 GL888413 EGI61325.1 GBBI01001032 JAC17680.1 KQ979308 KYN21974.1 KQ414663 KOC65440.1 LBMM01000796 KMQ97469.1 KQ434885 KZC10066.1 KQ971343 EFA03295.1 GL445250 EFN89984.1 GL440492 EFN65824.1 GEZM01078253 GEZM01078252 JAV62821.1 GECZ01028067 JAS41702.1 NNAY01004075 OXU18407.1 AAZX01014366 KZ288322 PBC28120.1 GECU01011935 JAS95771.1 GBYB01001023 JAG70790.1 KQ977085 KYN05916.1 KK107152 QOIP01000007 EZA57153.1 RLU20694.1 KQ762520 OAD55765.1 DS235124 EEB12243.1 GBHO01006416 GBRD01013866 GDHC01014428 JAG37188.1 JAG51960.1 JAQ04201.1 NEDP02005553 OWF38862.1 GGMR01011681 MBY24300.1 GGMS01001815 MBY71018.1 ABLF02033113 GFXV01007993 MBW19798.1 AK341378 BAH71785.1 GFPF01007664 MAA18810.1 GEDV01008764 JAP79793.1 GACK01006742 JAA58292.1 GDIP01225243 GDIP01225242 JAI98158.1 GEFH01003090 JAP65491.1 GFWV01002582 MAA27312.1 GBBM01001052 JAC34366.1 JO840646 AEO32263.1 GEFM01003153 JAP72643.1 GEDC01023657 JAS13641.1 GANP01000321 JAB84147.1 LJIG01022450 KRT80530.1 GACK01001058 JAA63976.1 JH817849 EKC30098.1 ABJB010081940 ABJB010688253 DS885859 EEC15238.1 APGK01029293 KB740731 KB632226 ENN79288.1 ERL90308.1 BT128199 AEE63160.1 AXCM01000997 GDIP01225241 JAI98160.1 GFDF01000777 JAV13307.1 GDIP01058857 JAM44858.1 GDIP01158330 LRGB01002167 JAJ65072.1 KZS08857.1 GDAI01000059 JAI17544.1 AJWK01022559 CH477186 EAT48991.1 AKHW03003562 KYO34237.1 GDIP01104139 JAL99575.1 UFQS01000611 UFQT01000611 SSX05366.1 SSX25727.1 GDIP01229535 JAI93866.1 JXUM01117830 KQ566123 KXJ70378.1 AAAB01008859 EAA07612.5 APCN01000591 BC053134 AAH53134.1 GDIP01122792 JAL80922.1 AMQN01010124 KB307090 ELT99216.1 GAPW01001555 JAC12043.1 EU142260 ABV60396.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000235965 UP000027135 UP000075809 UP000078541 UP000005205 UP000053105 UP000007755 UP000078492 UP000053825 UP000036403 UP000076502 UP000007266 UP000008237 UP000000311 UP000215335 UP000002358 UP000005203 UP000242457 UP000078542 UP000053097 UP000279307 UP000009046 UP000242188 UP000007819 UP000192223 UP000005408 UP000001555 UP000019118 UP000030742 UP000076408 UP000075883 UP000265040 UP000076858 UP000092461 UP000075900 UP000008820 UP000050525 UP000075920 UP000069940 UP000249989 UP000075881 UP000189705 UP000075885 UP000075902 UP000007062 UP000085678 UP000261520 UP000075903 UP000075840 UP000014760
UP000235965 UP000027135 UP000075809 UP000078541 UP000005205 UP000053105 UP000007755 UP000078492 UP000053825 UP000036403 UP000076502 UP000007266 UP000008237 UP000000311 UP000215335 UP000002358 UP000005203 UP000242457 UP000078542 UP000053097 UP000279307 UP000009046 UP000242188 UP000007819 UP000192223 UP000005408 UP000001555 UP000019118 UP000030742 UP000076408 UP000075883 UP000265040 UP000076858 UP000092461 UP000075900 UP000008820 UP000050525 UP000075920 UP000069940 UP000249989 UP000075881 UP000189705 UP000075885 UP000075902 UP000007062 UP000085678 UP000261520 UP000075903 UP000075840 UP000014760
Pfam
PF05291 Bystin
Interpro
IPR007955
Bystin
ProteinModelPortal
B9VTR7
H9JDY7
A0A2A4JSW8
A0A2H1VZC7
A0A194R8J0
A0A1E1WJA4
+ More
A0A194PQB0 S4PCD0 A0A212FDH3 A0A437BIY6 A0A2J7QLA3 A0A069DYY4 A0A067R4M0 A0A151X427 A0A224XQS3 A0A0P4VTY8 R4G3T9 A0A0V0G7S8 A0A195FSR5 A0A158NKZ3 A0A0N0U5Y7 F4WWT7 A0A023F8K2 A0A151JAB3 A0A0L7R3H7 A0A0J7L4N6 A0A154PDU5 D6WMH7 E2B315 E2AL69 A0A1Y1KQX7 A0A1B6EUR0 A0A232EJ63 K7IXM5 A0A087ZR91 A0A2A3E8T3 A0A1B6J988 A0A0C9PJ85 A0A151IMC5 A0A026WML5 A0A310SEA2 E0VFT7 A0A0A9Z1G7 A0A210PQY0 A0A2S2P4F3 A0A2S2PZU0 J9JM85 A0A2H8U2M6 C4WV65 A0A224YYH0 A0A131YKG8 L7M348 A0A0P4YMB0 A0A131XIW8 A0A293LEZ3 A0A1W4W8Y5 A0A023GKF5 G3MFJ5 A0A131XZI2 A0A1B6CJX0 V5I370 A0A0T6AZQ0 L7MIE1 K1Q864 B7Q8R6 N6TMN1 A0A182XV21 J3JY75 A0A182LUN6 A0A0N7ZPI7 A0A1L8E3L3 A0A0P5YEH7 A0A3Q1HNM2 A0A0P5DFC2 A0A0K8TT50 A0A1B0GK48 A0A182RVA5 Q0C738 A0A151NBS7 A0A0P5V7L6 A0A336MBH4 A0A182VRQ3 A0A0P4YL19 A0A182H887 A0A182JQM6 A0A1U7RWJ7 A0A182PQ05 A0A182TI11 A0A1S4EUN3 Q7QD29 A0A1S3K8C8 A0A3B3ZJC4 A0A2Y9D307 A0A182HTY1 Q7T3F8 A0A0P5TR74 R7U6A6 A0A023ES33 A8D881
A0A194PQB0 S4PCD0 A0A212FDH3 A0A437BIY6 A0A2J7QLA3 A0A069DYY4 A0A067R4M0 A0A151X427 A0A224XQS3 A0A0P4VTY8 R4G3T9 A0A0V0G7S8 A0A195FSR5 A0A158NKZ3 A0A0N0U5Y7 F4WWT7 A0A023F8K2 A0A151JAB3 A0A0L7R3H7 A0A0J7L4N6 A0A154PDU5 D6WMH7 E2B315 E2AL69 A0A1Y1KQX7 A0A1B6EUR0 A0A232EJ63 K7IXM5 A0A087ZR91 A0A2A3E8T3 A0A1B6J988 A0A0C9PJ85 A0A151IMC5 A0A026WML5 A0A310SEA2 E0VFT7 A0A0A9Z1G7 A0A210PQY0 A0A2S2P4F3 A0A2S2PZU0 J9JM85 A0A2H8U2M6 C4WV65 A0A224YYH0 A0A131YKG8 L7M348 A0A0P4YMB0 A0A131XIW8 A0A293LEZ3 A0A1W4W8Y5 A0A023GKF5 G3MFJ5 A0A131XZI2 A0A1B6CJX0 V5I370 A0A0T6AZQ0 L7MIE1 K1Q864 B7Q8R6 N6TMN1 A0A182XV21 J3JY75 A0A182LUN6 A0A0N7ZPI7 A0A1L8E3L3 A0A0P5YEH7 A0A3Q1HNM2 A0A0P5DFC2 A0A0K8TT50 A0A1B0GK48 A0A182RVA5 Q0C738 A0A151NBS7 A0A0P5V7L6 A0A336MBH4 A0A182VRQ3 A0A0P4YL19 A0A182H887 A0A182JQM6 A0A1U7RWJ7 A0A182PQ05 A0A182TI11 A0A1S4EUN3 Q7QD29 A0A1S3K8C8 A0A3B3ZJC4 A0A2Y9D307 A0A182HTY1 Q7T3F8 A0A0P5TR74 R7U6A6 A0A023ES33 A8D881
PDB
6G4W
E-value=1.42721e-129,
Score=1186
Ontologies
PANTHER
Topology
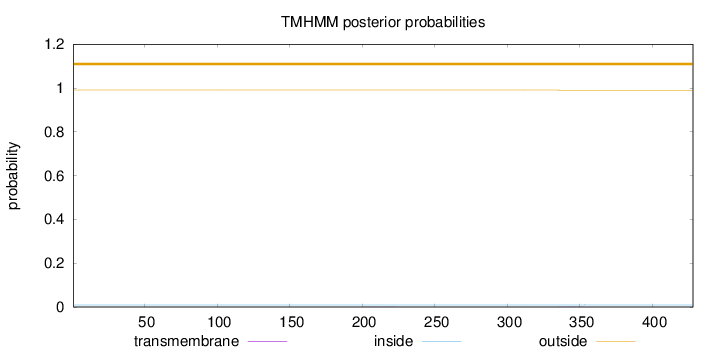
Length:
428
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01327
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00934
outside
1 - 428
Population Genetic Test Statistics
Pi
157.259355
Theta
132.214704
Tajima's D
0.08585
CLR
0.173426
CSRT
0.39718014099295
Interpretation
Uncertain
