Gene
KWMTBOMO08963
Pre Gene Modal
BGIBMGA007733
Annotation
hypothetical_protein_KGM_17464_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 2.161
Sequence
CDS
ATGTACAAGTATTCGGCTGTTCTAATACTGGCGCTGTTGGTTGCAACAGAGGGCCATGATTTTGTTGCCCAGAGCCAATTCCATTCTCAAGCCAGGAATGCCGTCACTCGTAACTGGGAAGGCCTTGGTCTAACAGCAGATATCAACGCATACGTCGACAGCATGATTGATACATTGGTTCCGTTCATGATAAAACATGACCTTGATCCTTTAAGTATTCCCGACATCGAAGAGACTTTTGAAGTTAGACCAGTCCTTATTACCTATAGAGCGAGTTTGGGTCTTACTAATGGCAAAATGAGCGGCTTAGTAAACGTGGCTCGTTCAGGAGATCAGTTGGTGCATTACTTCGCAAAGATGTTGCGTGCCCAGGCCAAATTTAAATTTACCGATCTTGAGTTCGATTTCGATTATCGCGTGAAAGTAATGAACATCGGACCTACTGGTCGCATCAGGGGTTCTTTGTCTCAATTCGTTATAAATGTTGATCTCATCATCGACTTCAACAACGATGAGATTCATCTTCAAGAGCTTTATCTAACTGACGTTGGTGTTATTCGAGTGAGCCTTTCTGGAAACATTCTCTACGATTGGTTGGTGACTCCTTTGGCAAATGTGTTTGTGCGTCTTTTCGATAATGTCATCATGGGATTCGTTGAAACCACTGTTAAAAGCATTGCTGGTTCCGCTATTGCTGATGTCAATGTCAACTTGAAACAAGTAATCCGCGATCTCGAAGCATTGAACAATTGA
Protein
MYKYSAVLILALLVATEGHDFVAQSQFHSQARNAVTRNWEGLGLTADINAYVDSMIDTLVPFMIKHDLDPLSIPDIEETFEVRPVLITYRASLGLTNGKMSGLVNVARSGDQLVHYFAKMLRAQAKFKFTDLEFDFDYRVKVMNIGPTGRIRGSLSQFVINVDLIIDFNNDEIHLQELYLTDVGVIRVSLSGNILYDWLVTPLANVFVRLFDNVIMGFVETTVKSIAGSAIADVNVNLKQVIRDLEALNN
Summary
Uniprot
H9JDY6
A0A2H1VXN4
A0A3S2NUC5
A0A2A4JSP4
A0A212EP06
A0A194R7T0
+ More
A0A194PKR1 A0A1J1IDS4 A0A182RZG2 A0A182QGH9 A0A084VYU9 A0A182MDM6 A0A182IX87 A0A182PS18 A0A2C9H8D7 A0A182HZA5 Q5TT57 A0A182KXH9 A0A182UW06 T1DPT4 A0A182FT14 A0A182GGD7 Q17F46 A0A1S4F574 D6WNT5 A0A1B0DDX1 A0A1W4WZK5 A0A139WHS5 A0A182NLG7 A0A2P8YU28 D6WUX7 A0A2P8YU10 A0A067RC34 V5GWS8 A0A0R3NJT6 A0A2J7PCM6 A0A0L7RII6 A0A0A9Y784 A0A0K8T030 A0A2J7RE71 A0A088AHX1 T1PCH7 A0A1Y1KYP0 A0A151JSA6 A0A1I8NUX6 A0A2J7QBY3 A0A2P8YU02 A0A2J7QLD0 A0A154P1V3 A0A151WJX9 A0A1B6DVX7 A0A195EW22 F4WVC0 A0A3L8DPY0 A0A2J7QBY0 A0A0N0BBP8 A0A0L0BMS6 A0A0C9R989 A0A2J7QBY7 A0A158NNU3 A0A195BS03 A0A1B6MHN9 A0A026WTI7 A0A310SBE7 A0A2J7QBX4 A0A1B6IX59 A0A1B6FYK3 A0A1I8IW02 A0A1I8I9N4
A0A194PKR1 A0A1J1IDS4 A0A182RZG2 A0A182QGH9 A0A084VYU9 A0A182MDM6 A0A182IX87 A0A182PS18 A0A2C9H8D7 A0A182HZA5 Q5TT57 A0A182KXH9 A0A182UW06 T1DPT4 A0A182FT14 A0A182GGD7 Q17F46 A0A1S4F574 D6WNT5 A0A1B0DDX1 A0A1W4WZK5 A0A139WHS5 A0A182NLG7 A0A2P8YU28 D6WUX7 A0A2P8YU10 A0A067RC34 V5GWS8 A0A0R3NJT6 A0A2J7PCM6 A0A0L7RII6 A0A0A9Y784 A0A0K8T030 A0A2J7RE71 A0A088AHX1 T1PCH7 A0A1Y1KYP0 A0A151JSA6 A0A1I8NUX6 A0A2J7QBY3 A0A2P8YU02 A0A2J7QLD0 A0A154P1V3 A0A151WJX9 A0A1B6DVX7 A0A195EW22 F4WVC0 A0A3L8DPY0 A0A2J7QBY0 A0A0N0BBP8 A0A0L0BMS6 A0A0C9R989 A0A2J7QBY7 A0A158NNU3 A0A195BS03 A0A1B6MHN9 A0A026WTI7 A0A310SBE7 A0A2J7QBX4 A0A1B6IX59 A0A1B6FYK3 A0A1I8IW02 A0A1I8I9N4
Pubmed
EMBL
BABH01002589
ODYU01005069
SOQ45583.1
RSAL01000187
RVE44790.1
NWSH01000740
+ More
PCG74473.1 AGBW02013568 OWR43206.1 KQ460615 KPJ13564.1 KQ459602 KPI93319.1 CVRI01000047 CRK98368.1 AXCN02000136 ATLV01018483 ATLV01018484 KE525234 KFB43143.1 AXCM01000605 AXCM01000606 APCN01005517 APCN01005518 AAAB01008888 EAL40593.2 GAMD01003069 JAA98521.1 JXUM01061579 JXUM01061580 KQ562158 KXJ76549.1 CH477276 EAT45137.1 KQ971342 EFA03851.1 AJVK01005493 AJVK01005494 KYB27550.1 PYGN01000359 PSN47753.1 KQ971357 EFA08505.1 PSN47749.1 KK852743 KDR17360.1 GALX01003743 JAB64723.1 CM000070 KRT01052.1 NEVH01027058 PNF14083.1 KQ414583 KOC70777.1 GBHO01028585 GBHO01016656 JAG15019.1 JAG26948.1 GBRD01007311 JAG58510.1 NEVH01005277 PNF39129.1 KA645648 AFP60277.1 GEZM01069769 JAV66512.1 KQ978538 KYN30253.1 NEVH01016293 PNF26091.1 PSN47733.1 NEVH01013250 PNF29368.1 KQ434786 KZC05110.1 KQ983031 KYQ48110.1 GEDC01007462 JAS29836.1 KQ981953 KYN32465.1 GL888384 EGI61886.1 QOIP01000006 RLU21959.1 PNF26093.1 KQ435969 KOX67861.1 JRES01001625 KNC21346.1 GBYB01009442 JAG79209.1 PNF26092.1 ADTU01021788 KQ976417 KYM89795.1 GEBQ01004578 JAT35399.1 KK107128 EZA58414.1 KQ775960 OAD52224.1 PNF26094.1 GECU01016192 JAS91514.1 GECZ01014512 JAS55257.1 NIVC01000591 PAA80271.1 NIVC01002236 PAA59740.1
PCG74473.1 AGBW02013568 OWR43206.1 KQ460615 KPJ13564.1 KQ459602 KPI93319.1 CVRI01000047 CRK98368.1 AXCN02000136 ATLV01018483 ATLV01018484 KE525234 KFB43143.1 AXCM01000605 AXCM01000606 APCN01005517 APCN01005518 AAAB01008888 EAL40593.2 GAMD01003069 JAA98521.1 JXUM01061579 JXUM01061580 KQ562158 KXJ76549.1 CH477276 EAT45137.1 KQ971342 EFA03851.1 AJVK01005493 AJVK01005494 KYB27550.1 PYGN01000359 PSN47753.1 KQ971357 EFA08505.1 PSN47749.1 KK852743 KDR17360.1 GALX01003743 JAB64723.1 CM000070 KRT01052.1 NEVH01027058 PNF14083.1 KQ414583 KOC70777.1 GBHO01028585 GBHO01016656 JAG15019.1 JAG26948.1 GBRD01007311 JAG58510.1 NEVH01005277 PNF39129.1 KA645648 AFP60277.1 GEZM01069769 JAV66512.1 KQ978538 KYN30253.1 NEVH01016293 PNF26091.1 PSN47733.1 NEVH01013250 PNF29368.1 KQ434786 KZC05110.1 KQ983031 KYQ48110.1 GEDC01007462 JAS29836.1 KQ981953 KYN32465.1 GL888384 EGI61886.1 QOIP01000006 RLU21959.1 PNF26093.1 KQ435969 KOX67861.1 JRES01001625 KNC21346.1 GBYB01009442 JAG79209.1 PNF26092.1 ADTU01021788 KQ976417 KYM89795.1 GEBQ01004578 JAT35399.1 KK107128 EZA58414.1 KQ775960 OAD52224.1 PNF26094.1 GECU01016192 JAS91514.1 GECZ01014512 JAS55257.1 NIVC01000591 PAA80271.1 NIVC01002236 PAA59740.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000183832 UP000075900 UP000075886 UP000030765 UP000075883 UP000075880 UP000075885 UP000076407 UP000075840 UP000007062 UP000075882 UP000075903 UP000069272 UP000069940 UP000249989 UP000008820 UP000007266 UP000092462 UP000192223 UP000075884 UP000245037 UP000027135 UP000001819 UP000235965 UP000053825 UP000005203 UP000078492 UP000095300 UP000076502 UP000075809 UP000078541 UP000007755 UP000279307 UP000053105 UP000037069 UP000005205 UP000078540 UP000053097 UP000095280 UP000215902
UP000183832 UP000075900 UP000075886 UP000030765 UP000075883 UP000075880 UP000075885 UP000076407 UP000075840 UP000007062 UP000075882 UP000075903 UP000069272 UP000069940 UP000249989 UP000008820 UP000007266 UP000092462 UP000192223 UP000075884 UP000245037 UP000027135 UP000001819 UP000235965 UP000053825 UP000005203 UP000078492 UP000095300 UP000076502 UP000075809 UP000078541 UP000007755 UP000279307 UP000053105 UP000037069 UP000005205 UP000078540 UP000053097 UP000095280 UP000215902
PRIDE
Interpro
SUPFAM
SSF55394
SSF55394
Gene 3D
ProteinModelPortal
H9JDY6
A0A2H1VXN4
A0A3S2NUC5
A0A2A4JSP4
A0A212EP06
A0A194R7T0
+ More
A0A194PKR1 A0A1J1IDS4 A0A182RZG2 A0A182QGH9 A0A084VYU9 A0A182MDM6 A0A182IX87 A0A182PS18 A0A2C9H8D7 A0A182HZA5 Q5TT57 A0A182KXH9 A0A182UW06 T1DPT4 A0A182FT14 A0A182GGD7 Q17F46 A0A1S4F574 D6WNT5 A0A1B0DDX1 A0A1W4WZK5 A0A139WHS5 A0A182NLG7 A0A2P8YU28 D6WUX7 A0A2P8YU10 A0A067RC34 V5GWS8 A0A0R3NJT6 A0A2J7PCM6 A0A0L7RII6 A0A0A9Y784 A0A0K8T030 A0A2J7RE71 A0A088AHX1 T1PCH7 A0A1Y1KYP0 A0A151JSA6 A0A1I8NUX6 A0A2J7QBY3 A0A2P8YU02 A0A2J7QLD0 A0A154P1V3 A0A151WJX9 A0A1B6DVX7 A0A195EW22 F4WVC0 A0A3L8DPY0 A0A2J7QBY0 A0A0N0BBP8 A0A0L0BMS6 A0A0C9R989 A0A2J7QBY7 A0A158NNU3 A0A195BS03 A0A1B6MHN9 A0A026WTI7 A0A310SBE7 A0A2J7QBX4 A0A1B6IX59 A0A1B6FYK3 A0A1I8IW02 A0A1I8I9N4
A0A194PKR1 A0A1J1IDS4 A0A182RZG2 A0A182QGH9 A0A084VYU9 A0A182MDM6 A0A182IX87 A0A182PS18 A0A2C9H8D7 A0A182HZA5 Q5TT57 A0A182KXH9 A0A182UW06 T1DPT4 A0A182FT14 A0A182GGD7 Q17F46 A0A1S4F574 D6WNT5 A0A1B0DDX1 A0A1W4WZK5 A0A139WHS5 A0A182NLG7 A0A2P8YU28 D6WUX7 A0A2P8YU10 A0A067RC34 V5GWS8 A0A0R3NJT6 A0A2J7PCM6 A0A0L7RII6 A0A0A9Y784 A0A0K8T030 A0A2J7RE71 A0A088AHX1 T1PCH7 A0A1Y1KYP0 A0A151JSA6 A0A1I8NUX6 A0A2J7QBY3 A0A2P8YU02 A0A2J7QLD0 A0A154P1V3 A0A151WJX9 A0A1B6DVX7 A0A195EW22 F4WVC0 A0A3L8DPY0 A0A2J7QBY0 A0A0N0BBP8 A0A0L0BMS6 A0A0C9R989 A0A2J7QBY7 A0A158NNU3 A0A195BS03 A0A1B6MHN9 A0A026WTI7 A0A310SBE7 A0A2J7QBX4 A0A1B6IX59 A0A1B6FYK3 A0A1I8IW02 A0A1I8I9N4
PDB
3UV1
E-value=0.000285196,
Score=102
Ontologies
Topology
Subcellular location
Nucleus
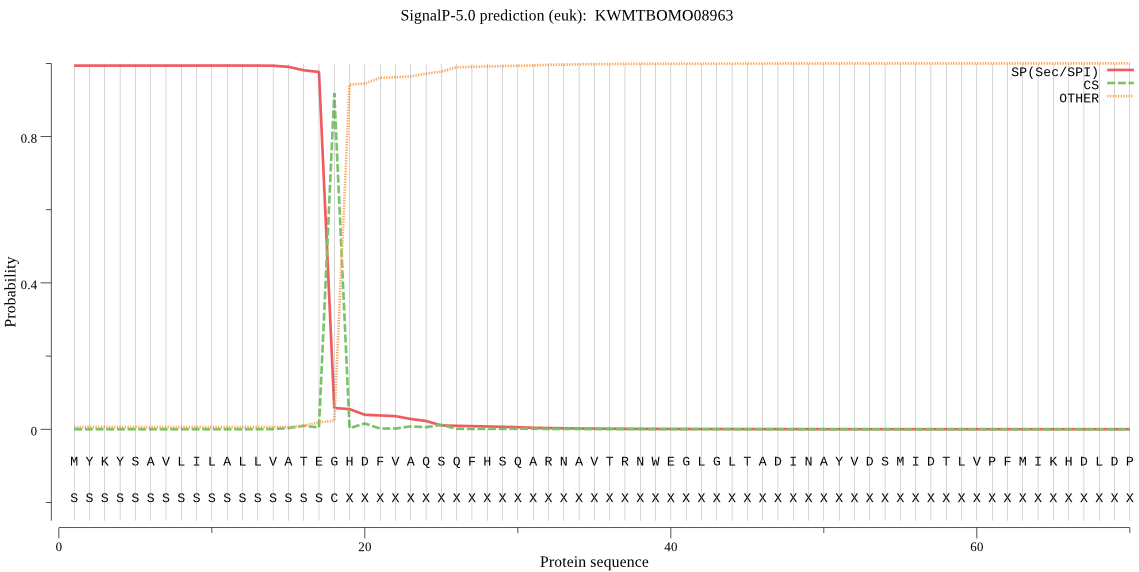
SignalP
Position: 1 - 18,
Likelihood: 0.993731
Length:
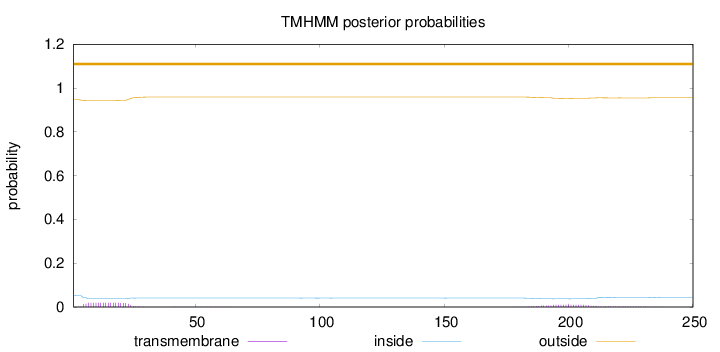
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.624900000000001
Exp number, first 60 AAs:
0.36984
Total prob of N-in:
0.05284
outside
1 - 250
Population Genetic Test Statistics
Pi
203.916977
Theta
169.82339
Tajima's D
0.744564
CLR
0.079189
CSRT
0.585770711464427
Interpretation
Uncertain
