Gene
KWMTBOMO08956 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007728
Annotation
PREDICTED:_cytosolic_non-specific_dipeptidase_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 4.118
Sequence
CDS
ATGGCAACGGAAAAAACATTGCCTGAAATTTTTAAATACGTAGATCAGAATAAAGATTCTTATAAGCAATTATTAAAAGAAGCAGTAGCCATTCCATCCGTTTCATGTGATGTGAAATATCGTGCGGATTGCATCCGTATGGTTCACTGGATGCAAGACAAATTGAAGGAAGTCGGAGCCACTACAGAGTTAAGGGATGTTGGCTTTCAAACAATAGATGGCAAAGACGTTCAACTGCCGCCAGTTTTAGTAGGCGTATTGGGTAATGATCCAAAAAAAAATACAGTTTGTATCTATGGTCATTTGGATGTACAACCTGCATTGAAATCTGATGGATGGGAGACTGAACCTTTTGAACTAGTTGAGCGCAATGAAAAATTATATGGAAGAGGTTCTACTGATGATAAAGGTCCTGTACTTGGTTGGCTGCATACCATCAATGCCTATAAGGGCACTGGTGCTGAGCTGCCTGTCAATTTGAAGTTCATTTTCGAATGTATGGAGGAATCTGGTTCCGAAGGGCTTGACAGCTTATTAATGGACAAATTGAAACCTGAGGGATTCTTTGATAGTGTAGACTATGTATGTATATCCGATAACTATTGGTTGGGAACCACGAAGCCATGTATCACTTATGGTTTACGCGGAATCAGCTACTATTTCCTAGAAGTGGAATGTGCAAAAATGGATCTTCACAGTGGAGTCTATGGTGGCACAGTTCATGAAGCCATGTCTGATTTAATCTACTTGATGAATACACTCGTGGACAAAGATGGGAAGATATTGGTGACTGACATATATAAATCGGTAGCTCCACTCACTGAAACCGAAACCAAGCTGTACACGACTATCGACTTTGATCCGGAAGCATACAGAAAGACAATTGGAGCATACAAACTTGCGCACAATGGCGTAAAAGAACAGTTGCTGATGCATCGTTGGAGATATCCCAGCCTATCTCTACACGGGATTGAAGGTGCCGCGTTCCAGCCTGGAGCAAAAACGGTGGTTCCTGGAAAAGTGATTGGAAAATTTTCAGTCAGGCTTGTTCCGAATCAAGAGCCGGAGGAAGTTGAGAAATTAGTCTTTGATTACATCAACAAGAAGTGGGCGGAGCGCGGGTCCCCGAACAAGATGAGCATCACGGCACAGAGCGGGCGCGCCTGGACCGAGAACCCGGACCATCCGCACTACCAGGCCGCTGCCCGCGCCACTAAGCTGATATATCAGACCGATCCGGACATGTCCCGCGAAGGAGGTTCGATCCCCGTCACGATCACTCTGCAGGAGGCGAGCGGCAAGAACGTGTTGCTGTTGCCCATGGGCGCCGGAGATGACATGGCGCACTCTCAAAACGAGAAGATTAACGTTCGCAACTACATCGAAGGCATAAAACTGTTTGCCGCCTACTTGTTCGAGGTCGGTAAACTCCCCAAGTAA
Protein
MATEKTLPEIFKYVDQNKDSYKQLLKEAVAIPSVSCDVKYRADCIRMVHWMQDKLKEVGATTELRDVGFQTIDGKDVQLPPVLVGVLGNDPKKNTVCIYGHLDVQPALKSDGWETEPFELVERNEKLYGRGSTDDKGPVLGWLHTINAYKGTGAELPVNLKFIFECMEESGSEGLDSLLMDKLKPEGFFDSVDYVCISDNYWLGTTKPCITYGLRGISYYFLEVECAKMDLHSGVYGGTVHEAMSDLIYLMNTLVDKDGKILVTDIYKSVAPLTETETKLYTTIDFDPEAYRKTIGAYKLAHNGVKEQLLMHRWRYPSLSLHGIEGAAFQPGAKTVVPGKVIGKFSVRLVPNQEPEEVEKLVFDYINKKWAERGSPNKMSITAQSGRAWTENPDHPHYQAAARATKLIYQTDPDMSREGGSIPVTITLQEASGKNVLLLPMGAGDDMAHSQNEKINVRNYIEGIKLFAAYLFEVGKLPK
Summary
Cofactor
Mn(2+)
Uniprot
H9JDY1
A0A194R8H5
A0A194PIR4
A0A3S2NRC9
A0A2A4JS91
S4PF17
+ More
A0A212FGT1 A0A1J1JBB1 A0A336L7Z7 N6TE70 A0A154PLI6 K7J647 U5EU96 A0A232EVH4 A0A023ESZ6 A0A1B6LJX5 A0A182G359 N6UBK2 B0XCT6 Q17KV3 A0A182FFV8 A0A2M4A5H5 A0A2M4BM01 A0A411G7I2 A0A2M4BLR0 A0A2M4A4T0 A0A310SGQ9 A0A2M3ZH36 A0A1W4XA82 T1DNF9 A0A2M3ZH25 W5JEW2 A0A2M4BLN5 A0A1Q3F977 A0A182IP61 D6WGK3 A0A182X6C9 A0A182VHH7 A0A182KRS9 A0A182I3N5 A0A1B6HGQ3 E2B4G3 A0A084VX00 A0A182WIQ5 A0A151J029 Q7QJQ9 V5I9Z3 A0A182MHM3 A0A195CUQ0 A0A182RFM3 A0A182P3L2 A0A182Q4H5 E9IN77 A0A1I8NDN8 A0A182JX13 A0A026WJS4 A0A158NQ49 A0A0J7KP64 A0A195FPB8 E2A1T0 A0A182XZ81 A0A182NHM6 A0A1L8EFI5 A0A034WDN8 J3JUI2 D3TRW2 F4WMR8 A0A1B0AM21 A0A1I8PKV4 W8C5B6 A0A195BJ63 A0A0K8R3X9 A0A1A9WI90 A0A1L8DQS9 A0A0A1WGN5 B4NZC0 A0A0L0CDT3 A0A1Y1N383 B7PIM5 A0A0J9R6I9 B4II02 A0A2A3E0C0 V9IGN0 Q8MT58 A0A1B0CCN4 B3N3J1 A0A224YUH8 A0A0P4W9P8 L7M7B2 A0A1S4MXZ6 A0A1L8DR24 A0A131YQQ5 B4MFS0 A0A088ADU9 A0A0K8WIA1 A0A0L7R996 A0A3B0J3Q3 V5G6F8 A0A2J7R470
A0A212FGT1 A0A1J1JBB1 A0A336L7Z7 N6TE70 A0A154PLI6 K7J647 U5EU96 A0A232EVH4 A0A023ESZ6 A0A1B6LJX5 A0A182G359 N6UBK2 B0XCT6 Q17KV3 A0A182FFV8 A0A2M4A5H5 A0A2M4BM01 A0A411G7I2 A0A2M4BLR0 A0A2M4A4T0 A0A310SGQ9 A0A2M3ZH36 A0A1W4XA82 T1DNF9 A0A2M3ZH25 W5JEW2 A0A2M4BLN5 A0A1Q3F977 A0A182IP61 D6WGK3 A0A182X6C9 A0A182VHH7 A0A182KRS9 A0A182I3N5 A0A1B6HGQ3 E2B4G3 A0A084VX00 A0A182WIQ5 A0A151J029 Q7QJQ9 V5I9Z3 A0A182MHM3 A0A195CUQ0 A0A182RFM3 A0A182P3L2 A0A182Q4H5 E9IN77 A0A1I8NDN8 A0A182JX13 A0A026WJS4 A0A158NQ49 A0A0J7KP64 A0A195FPB8 E2A1T0 A0A182XZ81 A0A182NHM6 A0A1L8EFI5 A0A034WDN8 J3JUI2 D3TRW2 F4WMR8 A0A1B0AM21 A0A1I8PKV4 W8C5B6 A0A195BJ63 A0A0K8R3X9 A0A1A9WI90 A0A1L8DQS9 A0A0A1WGN5 B4NZC0 A0A0L0CDT3 A0A1Y1N383 B7PIM5 A0A0J9R6I9 B4II02 A0A2A3E0C0 V9IGN0 Q8MT58 A0A1B0CCN4 B3N3J1 A0A224YUH8 A0A0P4W9P8 L7M7B2 A0A1S4MXZ6 A0A1L8DR24 A0A131YQQ5 B4MFS0 A0A088ADU9 A0A0K8WIA1 A0A0L7R996 A0A3B0J3Q3 V5G6F8 A0A2J7R470
Pubmed
19121390
26354079
23622113
22118469
23537049
20075255
+ More
28648823 24945155 26483478 17510324 20920257 23761445 18362917 19820115 20966253 20798317 24438588 12364791 14747013 17210077 21282665 25315136 24508170 30249741 21347285 25244985 25348373 22516182 20353571 21719571 24495485 25830018 17994087 17550304 26108605 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 28797301 25576852 20566863 26830274
28648823 24945155 26483478 17510324 20920257 23761445 18362917 19820115 20966253 20798317 24438588 12364791 14747013 17210077 21282665 25315136 24508170 30249741 21347285 25244985 25348373 22516182 20353571 21719571 24495485 25830018 17994087 17550304 26108605 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 28797301 25576852 20566863 26830274
EMBL
BABH01002596
BABH01002597
KQ460615
KPJ13555.1
KQ459602
KPI93311.1
+ More
RSAL01000315 RVE42615.1 NWSH01000740 PCG74474.1 GAIX01004247 JAA88313.1 AGBW02008599 OWR52934.1 CVRI01000075 CRL08345.1 UFQS01002047 UFQT01002047 UFQT01002956 SSX13091.1 SSX32531.1 SSX34342.1 APGK01041353 KB740993 ENN76033.1 KQ434938 KZC12090.1 GANO01001582 JAB58289.1 NNAY01002011 OXU22315.1 GAPW01001181 JAC12417.1 GEBQ01021778 GEBQ01016063 GEBQ01004599 JAT18199.1 JAT23914.1 JAT35378.1 JXUM01040552 KQ561249 KXJ79229.1 ENN76032.1 DS232718 EDS45103.1 CH477221 EAT47301.1 GGFK01002680 MBW36001.1 GGFJ01004832 MBW53973.1 MH366060 QBB01869.1 GGFJ01004831 MBW53972.1 GGFK01002486 MBW35807.1 KQ769940 OAD52750.1 GGFM01007044 MBW27795.1 GAMD01002920 JAA98670.1 GGFM01007050 MBW27801.1 ADMH02001692 ETN61410.1 GGFJ01004836 MBW53977.1 GFDL01010968 JAV24077.1 KQ971321 EFA00576.1 APCN01000215 GECU01033819 JAS73887.1 GL445556 EFN89424.1 ATLV01017810 KE525192 KFB42494.1 KQ980658 KYN14772.1 AAAB01008807 EAA03960.4 GALX01002085 JAB66381.1 AXCM01000215 KQ977279 KYN04247.1 AXCN02000593 GL764290 EFZ17970.1 KK107167 QOIP01000010 EZA56255.1 RLU17744.1 ADTU01022963 LBMM01004716 KMQ92148.1 KQ981382 KYN42157.1 GL435798 EFN72602.1 GFDG01001312 JAV17487.1 GAKP01006500 JAC52452.1 BT126896 AEE61858.1 EZ424164 ADD20440.1 GL888218 EGI64545.1 JXJN01000314 GAMC01004970 GAMC01004969 JAC01587.1 KQ976464 KYM84409.1 GADI01007948 JAA65860.1 GFDF01005258 JAV08826.1 GBXI01016108 JAC98183.1 CM000157 EDW89833.1 JRES01000644 KNC29654.1 GEZM01013821 JAV92312.1 ABJB010679286 DS720013 EEC06447.1 CM002911 KMY91641.1 CH480841 EDW49528.1 KZ288497 PBC25178.1 JR042559 AEY59636.1 AE013599 AY118368 AAF57318.2 AAM48397.1 AJWK01006927 AJWK01006928 CH954177 EDV59873.1 EDV59874.1 GFPF01007105 MAA18251.1 GDRN01070213 JAI63918.1 GACK01004863 JAA60171.1 AAZO01006524 GFDF01005257 JAV08827.1 GEDV01007747 JAP80810.1 CH940667 EDW57241.2 GDHF01001436 JAI50878.1 KQ414627 KOC67450.1 OUUW01000001 SPP75955.1 GALX01002801 JAB65665.1 NEVH01007815 PNF35628.1
RSAL01000315 RVE42615.1 NWSH01000740 PCG74474.1 GAIX01004247 JAA88313.1 AGBW02008599 OWR52934.1 CVRI01000075 CRL08345.1 UFQS01002047 UFQT01002047 UFQT01002956 SSX13091.1 SSX32531.1 SSX34342.1 APGK01041353 KB740993 ENN76033.1 KQ434938 KZC12090.1 GANO01001582 JAB58289.1 NNAY01002011 OXU22315.1 GAPW01001181 JAC12417.1 GEBQ01021778 GEBQ01016063 GEBQ01004599 JAT18199.1 JAT23914.1 JAT35378.1 JXUM01040552 KQ561249 KXJ79229.1 ENN76032.1 DS232718 EDS45103.1 CH477221 EAT47301.1 GGFK01002680 MBW36001.1 GGFJ01004832 MBW53973.1 MH366060 QBB01869.1 GGFJ01004831 MBW53972.1 GGFK01002486 MBW35807.1 KQ769940 OAD52750.1 GGFM01007044 MBW27795.1 GAMD01002920 JAA98670.1 GGFM01007050 MBW27801.1 ADMH02001692 ETN61410.1 GGFJ01004836 MBW53977.1 GFDL01010968 JAV24077.1 KQ971321 EFA00576.1 APCN01000215 GECU01033819 JAS73887.1 GL445556 EFN89424.1 ATLV01017810 KE525192 KFB42494.1 KQ980658 KYN14772.1 AAAB01008807 EAA03960.4 GALX01002085 JAB66381.1 AXCM01000215 KQ977279 KYN04247.1 AXCN02000593 GL764290 EFZ17970.1 KK107167 QOIP01000010 EZA56255.1 RLU17744.1 ADTU01022963 LBMM01004716 KMQ92148.1 KQ981382 KYN42157.1 GL435798 EFN72602.1 GFDG01001312 JAV17487.1 GAKP01006500 JAC52452.1 BT126896 AEE61858.1 EZ424164 ADD20440.1 GL888218 EGI64545.1 JXJN01000314 GAMC01004970 GAMC01004969 JAC01587.1 KQ976464 KYM84409.1 GADI01007948 JAA65860.1 GFDF01005258 JAV08826.1 GBXI01016108 JAC98183.1 CM000157 EDW89833.1 JRES01000644 KNC29654.1 GEZM01013821 JAV92312.1 ABJB010679286 DS720013 EEC06447.1 CM002911 KMY91641.1 CH480841 EDW49528.1 KZ288497 PBC25178.1 JR042559 AEY59636.1 AE013599 AY118368 AAF57318.2 AAM48397.1 AJWK01006927 AJWK01006928 CH954177 EDV59873.1 EDV59874.1 GFPF01007105 MAA18251.1 GDRN01070213 JAI63918.1 GACK01004863 JAA60171.1 AAZO01006524 GFDF01005257 JAV08827.1 GEDV01007747 JAP80810.1 CH940667 EDW57241.2 GDHF01001436 JAI50878.1 KQ414627 KOC67450.1 OUUW01000001 SPP75955.1 GALX01002801 JAB65665.1 NEVH01007815 PNF35628.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000218220
UP000007151
+ More
UP000183832 UP000019118 UP000076502 UP000002358 UP000215335 UP000069940 UP000249989 UP000002320 UP000008820 UP000069272 UP000192223 UP000000673 UP000075880 UP000007266 UP000076407 UP000075903 UP000075882 UP000075840 UP000008237 UP000030765 UP000075920 UP000078492 UP000007062 UP000075883 UP000078542 UP000075900 UP000075885 UP000075886 UP000095301 UP000075881 UP000053097 UP000279307 UP000005205 UP000036403 UP000078541 UP000000311 UP000076408 UP000075884 UP000007755 UP000092460 UP000095300 UP000078540 UP000091820 UP000002282 UP000037069 UP000001555 UP000001292 UP000242457 UP000000803 UP000092461 UP000008711 UP000008792 UP000005203 UP000053825 UP000268350 UP000235965
UP000183832 UP000019118 UP000076502 UP000002358 UP000215335 UP000069940 UP000249989 UP000002320 UP000008820 UP000069272 UP000192223 UP000000673 UP000075880 UP000007266 UP000076407 UP000075903 UP000075882 UP000075840 UP000008237 UP000030765 UP000075920 UP000078492 UP000007062 UP000075883 UP000078542 UP000075900 UP000075885 UP000075886 UP000095301 UP000075881 UP000053097 UP000279307 UP000005205 UP000036403 UP000078541 UP000000311 UP000076408 UP000075884 UP000007755 UP000092460 UP000095300 UP000078540 UP000091820 UP000002282 UP000037069 UP000001555 UP000001292 UP000242457 UP000000803 UP000092461 UP000008711 UP000008792 UP000005203 UP000053825 UP000268350 UP000235965
PRIDE
Pfam
Interpro
IPR017153
CNDP/DUG1
+ More
IPR001261 ArgE/DapE_CS
IPR011650 Peptidase_M20_dimer
IPR002933 Peptidase_M20
IPR036397 RNaseH_sf
IPR008936 Rho_GTPase_activation_prot
IPR000198 RhoGAP_dom
IPR000626 Ubiquitin_dom
IPR021925 BAG6
IPR029071 Ubiquitin-like_domsf
IPR005079 Peptidase_C45
IPR018253 DnaJ_domain_CS
IPR036869 J_dom_sf
IPR001623 DnaJ_domain
IPR017149 GSH_degradosome_Dug2
IPR001261 ArgE/DapE_CS
IPR011650 Peptidase_M20_dimer
IPR002933 Peptidase_M20
IPR036397 RNaseH_sf
IPR008936 Rho_GTPase_activation_prot
IPR000198 RhoGAP_dom
IPR000626 Ubiquitin_dom
IPR021925 BAG6
IPR029071 Ubiquitin-like_domsf
IPR005079 Peptidase_C45
IPR018253 DnaJ_domain_CS
IPR036869 J_dom_sf
IPR001623 DnaJ_domain
IPR017149 GSH_degradosome_Dug2
Gene 3D
ProteinModelPortal
H9JDY1
A0A194R8H5
A0A194PIR4
A0A3S2NRC9
A0A2A4JS91
S4PF17
+ More
A0A212FGT1 A0A1J1JBB1 A0A336L7Z7 N6TE70 A0A154PLI6 K7J647 U5EU96 A0A232EVH4 A0A023ESZ6 A0A1B6LJX5 A0A182G359 N6UBK2 B0XCT6 Q17KV3 A0A182FFV8 A0A2M4A5H5 A0A2M4BM01 A0A411G7I2 A0A2M4BLR0 A0A2M4A4T0 A0A310SGQ9 A0A2M3ZH36 A0A1W4XA82 T1DNF9 A0A2M3ZH25 W5JEW2 A0A2M4BLN5 A0A1Q3F977 A0A182IP61 D6WGK3 A0A182X6C9 A0A182VHH7 A0A182KRS9 A0A182I3N5 A0A1B6HGQ3 E2B4G3 A0A084VX00 A0A182WIQ5 A0A151J029 Q7QJQ9 V5I9Z3 A0A182MHM3 A0A195CUQ0 A0A182RFM3 A0A182P3L2 A0A182Q4H5 E9IN77 A0A1I8NDN8 A0A182JX13 A0A026WJS4 A0A158NQ49 A0A0J7KP64 A0A195FPB8 E2A1T0 A0A182XZ81 A0A182NHM6 A0A1L8EFI5 A0A034WDN8 J3JUI2 D3TRW2 F4WMR8 A0A1B0AM21 A0A1I8PKV4 W8C5B6 A0A195BJ63 A0A0K8R3X9 A0A1A9WI90 A0A1L8DQS9 A0A0A1WGN5 B4NZC0 A0A0L0CDT3 A0A1Y1N383 B7PIM5 A0A0J9R6I9 B4II02 A0A2A3E0C0 V9IGN0 Q8MT58 A0A1B0CCN4 B3N3J1 A0A224YUH8 A0A0P4W9P8 L7M7B2 A0A1S4MXZ6 A0A1L8DR24 A0A131YQQ5 B4MFS0 A0A088ADU9 A0A0K8WIA1 A0A0L7R996 A0A3B0J3Q3 V5G6F8 A0A2J7R470
A0A212FGT1 A0A1J1JBB1 A0A336L7Z7 N6TE70 A0A154PLI6 K7J647 U5EU96 A0A232EVH4 A0A023ESZ6 A0A1B6LJX5 A0A182G359 N6UBK2 B0XCT6 Q17KV3 A0A182FFV8 A0A2M4A5H5 A0A2M4BM01 A0A411G7I2 A0A2M4BLR0 A0A2M4A4T0 A0A310SGQ9 A0A2M3ZH36 A0A1W4XA82 T1DNF9 A0A2M3ZH25 W5JEW2 A0A2M4BLN5 A0A1Q3F977 A0A182IP61 D6WGK3 A0A182X6C9 A0A182VHH7 A0A182KRS9 A0A182I3N5 A0A1B6HGQ3 E2B4G3 A0A084VX00 A0A182WIQ5 A0A151J029 Q7QJQ9 V5I9Z3 A0A182MHM3 A0A195CUQ0 A0A182RFM3 A0A182P3L2 A0A182Q4H5 E9IN77 A0A1I8NDN8 A0A182JX13 A0A026WJS4 A0A158NQ49 A0A0J7KP64 A0A195FPB8 E2A1T0 A0A182XZ81 A0A182NHM6 A0A1L8EFI5 A0A034WDN8 J3JUI2 D3TRW2 F4WMR8 A0A1B0AM21 A0A1I8PKV4 W8C5B6 A0A195BJ63 A0A0K8R3X9 A0A1A9WI90 A0A1L8DQS9 A0A0A1WGN5 B4NZC0 A0A0L0CDT3 A0A1Y1N383 B7PIM5 A0A0J9R6I9 B4II02 A0A2A3E0C0 V9IGN0 Q8MT58 A0A1B0CCN4 B3N3J1 A0A224YUH8 A0A0P4W9P8 L7M7B2 A0A1S4MXZ6 A0A1L8DR24 A0A131YQQ5 B4MFS0 A0A088ADU9 A0A0K8WIA1 A0A0L7R996 A0A3B0J3Q3 V5G6F8 A0A2J7R470
PDB
2ZOG
E-value=6.83284e-158,
Score=1430
Ontologies
PATHWAY
GO
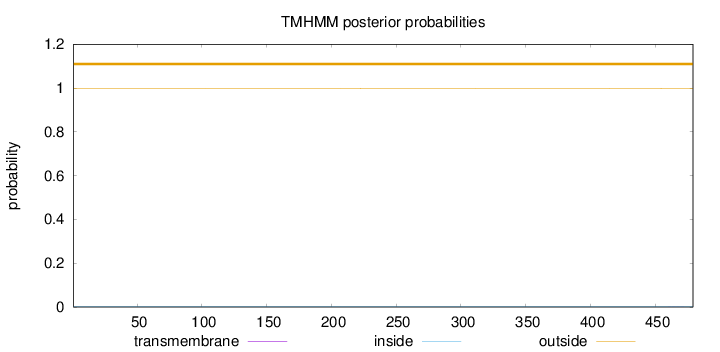
Topology
Length:
479
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00355
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00245
outside
1 - 479
Population Genetic Test Statistics
Pi
275.374759
Theta
172.181209
Tajima's D
1.580979
CLR
0.61927
CSRT
0.810259487025649
Interpretation
Uncertain
