Gene
KWMTBOMO08955 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007727
Annotation
PREDICTED:_nicastrin_[Bombyx_mori]
Full name
Nicastrin
Location in the cell
PlasmaMembrane Reliability : 4.482
Sequence
CDS
ATGGCTGTTGTCAGCACATCAAAATTTGGCCTAGTAGAGTTTCTCATAGATCATCCAACAAATATTGCAGGAGTACTTACATATGAAAATGCTTCTGATATCCCCTCATTTTTTACACATGACACAAGATGTCCAAATGAGTTTTCATCAGCAGATGGAAACCAGTGTTCATCTTCAAAGGAAGGTGGTGTTGTGTGGAATGAAAAAGGCACAGGTTTATTAAAGAGAGAAATACCATTTCCAATATTTTTCATACCAGAATCAAGATATGAAGAAATTGAAAAAATCAGTCATTGTTTCGGAAGATTCAATTACGATAAAAACAACCAAAGGGGTCGGTCGCTATGCTCAATACAGTTGAATTCCTTCATGTTTGCTGCAATAAATACCGAAGTTTGCATGAGAAGGTCAGCTACCTCAACGATAGTAACAACAACGAAAGTGTGCGATCCGCTTGGCGATCAAAACGTTTATTATTCCCTTTTCCCTAGAACGAAAGAATCTAAGAAGCAGTCCATCACGTTGGTGACTGCAAGGATCGATTCAGCTTCTTTATTTGATGGGGTGTCTCCGGGTGCTGCTAGTTCTGTCGTCGGCCTAGTCACGCTGATATCAGCAGCCGCCGCATTAGCTCAGCTGATTCCAGTTTCTAAATCGAATCTATACGAGACGAATATATTATGGACGTTATTCAACGGGGAAGCATTTGATTATATTGGTTCACAGCGAGTTGCCTACGACATAAAGCGCGGGGTGTGGCCGCCGAATGCACCGCTGGCACCGACCGATGTAAAACTACACGTTGAAATCGGCCAGATCGGTGGATCCTTATCAGATTATGTGAACGAAAATACATGGCCCTTGAACGCTTTTGTACCTACCCTCGCGCCATCAACAATAAATGACGGTGATAATGATATACAAATCTTCCTGAAACAATTGTCGAATGCATATAACATGACAATACCCGGAATCTTTAATGGCTCTCTGCCACCGTCATCTATACATTCATTCAGAAGAATATTATCCAATGTGACAGAGAGCGGTGAAATGCTTGAAATGTTATTAGTTGATCACAAAGAGACCTTCACGAATGTTTTCTACAACTCTGCTTTGGACGACAGTGACAAAATCGCTTATGTCTATAAAAACATCAGTATGATCAACAATGTTACATTTATATCCACAGCTGAACTCATAGCAGATGGTTTGATGAAAGAAACTGACACTCAAGTAAAGATATCTCGGTTGGCGACGGCTTTAGCTCATGCACTATATCAAAGAGTTGTCGGTGAGGCGTACACTGGTAATATTACAGTGTCCGCACATTTGGTAGATGAAATGCTGTACTGTTTCCTGAAGAACGAGACGTGCAAGTTGTTATCGGCTGCGGAGTTTGGCGGCAACGAGGGGCCGCGGGAGGGTGCAGGGCCGGCGCCGCTGTACGTGGGCGTGACCTCCTGGAACACGGTGGCGCCCGTGTTCGCCGGACACCTGCTGGCCTTGCTTACCGGCACGCAACTCTCGCTCAACAAATCACAGTGCGAAACGAAACCAGAATCGGGATATACAAGGCTCTGGTTAAAAGGATGGAATCACAATGGCGTTTGCATCGAGACCACAATGAACTTTAGTCAAGCACTCAGTCCAGCTTTTATAATATCAGATTATGATTTTAAATCGGGTACGTATTCTACGTGGACCGAGTCAGTTTGGCAAACAATGTGGGCCCGAGTGTTCGTGTCGGCGAGCGGCGGCAGCGCGTGCGTGGCGGCCGCGACCGGTGCCGTGTCAACCCTTATTGCGGCCCTGCTCACGTTTTGGCTACGCAGACACTCATCACTCATATTTGTCGATCCGCCGTCAGCAACCATCGTTGTTGACGATGCTGCTTCCGGAATTTTACGTTCTGTGAACTGTTAG
Protein
MAVVSTSKFGLVEFLIDHPTNIAGVLTYENASDIPSFFTHDTRCPNEFSSADGNQCSSSKEGGVVWNEKGTGLLKREIPFPIFFIPESRYEEIEKISHCFGRFNYDKNNQRGRSLCSIQLNSFMFAAINTEVCMRRSATSTIVTTTKVCDPLGDQNVYYSLFPRTKESKKQSITLVTARIDSASLFDGVSPGAASSVVGLVTLISAAAALAQLIPVSKSNLYETNILWTLFNGEAFDYIGSQRVAYDIKRGVWPPNAPLAPTDVKLHVEIGQIGGSLSDYVNENTWPLNAFVPTLAPSTINDGDNDIQIFLKQLSNAYNMTIPGIFNGSLPPSSIHSFRRILSNVTESGEMLEMLLVDHKETFTNVFYNSALDDSDKIAYVYKNISMINNVTFISTAELIADGLMKETDTQVKISRLATALAHALYQRVVGEAYTGNITVSAHLVDEMLYCFLKNETCKLLSAAEFGGNEGPREGAGPAPLYVGVTSWNTVAPVFAGHLLALLTGTQLSLNKSQCETKPESGYTRLWLKGWNHNGVCIETTMNFSQALSPAFIISDYDFKSGTYSTWTESVWQTMWARVFVSASGGSACVAAATGAVSTLIAALLTFWLRRHSSLIFVDPPSATIVVDDAASGILRSVNC
Summary
Description
Essential subunit of the gamma-secretase complex, an endoprotease complex that catalyzes the intramembrane cleavage of integral membrane proteins such as Notch. It probably represents a stabilizing cofactor required for the assembly of the gamma-secretase complex.
Subunit
Component of the gamma-secretase complex, a complex composed of a presenilin (Psn) homodimer, nicastrin (Nct), Aph-1 and Pen-2.
Similarity
Belongs to the nicastrin family.
Keywords
Complete proteome
Glycoprotein
Membrane
Notch signaling pathway
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Nicastrin
Uniprot
H9JDY0
A0A2H1WCH3
S4PFY1
A0A2A4JR01
A0A437AWS1
A0A212FGR9
+ More
A0A194PQA0 A0A194R7R5 A0A1E1WBR6 B3LV10 A0A1Q3FCX7 A0A3B0JCQ0 A0A1L8DM59 A0A1Y1L8T5 U5EXI1 B0W373 A0A0L7QWU1 A0A154PEW5 D2A5N1 B4QTU0 Q29CG8 A0A1B6JLP8 A0A067QWF2 A0A1B0GHV0 E1JIV8 Q9VC27 Q7KS07 A0A0B4KHG2 B4GN29 B4HHL8 Q17CA6 A0A1W4VR20 A0A1W4W349 A0A1S4F865 A0A182H1V0 A0A182GLG6 B3P6X4 B4PV32 A0A1B6M4G2 A0A1B6KU33 A0A195AUS2 A0A0N0U3I5 E2AJI0 A0A2M4BF61 A0A158NHP2 A0A182SDC3 A0A195E285 A0A1J1J526 B4NFN8 A0A2M4BF39 A0A1Y9HAG1 A0A2M4BFD8 W5JFV8 A0A151XFD1 F4WVP9 A0A182JJR2 A0A1Y9H2V6 A0A240PL32 A0A195FWU1 A0A0C9Q2S0 A0A182Y3U6 A0A195D4A0 A0A182U8L9 E2BHJ1 A0A1I8JSS5 A0A240PKR8 A0A1Y9IVQ9 A0A0M4EHB4 B4K4I5 A0A0Q9WXI5 A0A232F0X7 A0A2M3ZEC1 B4LVZ2 A0A0Q9WP97 A0A1D2MFT3 A0A0A1XBN2 A0A1I8JVR0 A0A1I8JTD9 T1HZX0 A0A182UP50 A0A2M4A8B1 K7ISR9 A0A2M4A852 A0A2M4A7P4 A0A336KLF8 A0A088AD09 A0A1I8JUS5 A0A182MPD3 A0A2A3EKB5 Q7PY38 A0A1S4GCU3 A0A084VZE3 W8C6X3 W8BAG5 A0A1B6EBK1 A0A0K8UPS5 A0A0K8V810 A0A2M3Z3F3 A0A0K8V067 A0A182LA47
A0A194PQA0 A0A194R7R5 A0A1E1WBR6 B3LV10 A0A1Q3FCX7 A0A3B0JCQ0 A0A1L8DM59 A0A1Y1L8T5 U5EXI1 B0W373 A0A0L7QWU1 A0A154PEW5 D2A5N1 B4QTU0 Q29CG8 A0A1B6JLP8 A0A067QWF2 A0A1B0GHV0 E1JIV8 Q9VC27 Q7KS07 A0A0B4KHG2 B4GN29 B4HHL8 Q17CA6 A0A1W4VR20 A0A1W4W349 A0A1S4F865 A0A182H1V0 A0A182GLG6 B3P6X4 B4PV32 A0A1B6M4G2 A0A1B6KU33 A0A195AUS2 A0A0N0U3I5 E2AJI0 A0A2M4BF61 A0A158NHP2 A0A182SDC3 A0A195E285 A0A1J1J526 B4NFN8 A0A2M4BF39 A0A1Y9HAG1 A0A2M4BFD8 W5JFV8 A0A151XFD1 F4WVP9 A0A182JJR2 A0A1Y9H2V6 A0A240PL32 A0A195FWU1 A0A0C9Q2S0 A0A182Y3U6 A0A195D4A0 A0A182U8L9 E2BHJ1 A0A1I8JSS5 A0A240PKR8 A0A1Y9IVQ9 A0A0M4EHB4 B4K4I5 A0A0Q9WXI5 A0A232F0X7 A0A2M3ZEC1 B4LVZ2 A0A0Q9WP97 A0A1D2MFT3 A0A0A1XBN2 A0A1I8JVR0 A0A1I8JTD9 T1HZX0 A0A182UP50 A0A2M4A8B1 K7ISR9 A0A2M4A852 A0A2M4A7P4 A0A336KLF8 A0A088AD09 A0A1I8JUS5 A0A182MPD3 A0A2A3EKB5 Q7PY38 A0A1S4GCU3 A0A084VZE3 W8C6X3 W8BAG5 A0A1B6EBK1 A0A0K8UPS5 A0A0K8V810 A0A2M3Z3F3 A0A0K8V067 A0A182LA47
Pubmed
19121390
23622113
22118469
26354079
17994087
28004739
+ More
18362917 19820115 15632085 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10993067 11781576 11782315 11782316 12660785 17510324 26483478 17550304 20798317 21347285 20920257 23761445 21719571 25244985 28648823 27289101 25830018 20075255 12364791 24438588 24495485 20966253
18362917 19820115 15632085 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10993067 11781576 11782315 11782316 12660785 17510324 26483478 17550304 20798317 21347285 20920257 23761445 21719571 25244985 28648823 27289101 25830018 20075255 12364791 24438588 24495485 20966253
EMBL
BABH01002597
BABH01002598
ODYU01007669
SOQ50656.1
GAIX01003847
JAA88713.1
+ More
NWSH01000740 PCG74475.1 RSAL01000315 RVE42617.1 AGBW02008599 OWR52935.1 KQ459602 KPI93310.1 KQ460615 KPJ13554.1 GDQN01006669 JAT84385.1 CH902617 EDV42482.1 GFDL01009692 JAV25353.1 OUUW01000005 SPP80154.1 GFDF01006664 JAV07420.1 GEZM01062164 JAV69994.1 GANO01000942 JAB58929.1 DS231831 EDS31216.1 KQ414706 KOC63093.1 KQ434889 KZC10347.1 KQ971345 EFA05049.2 CM000364 EDX14294.1 CM000070 EAL26678.4 GECU01007601 JAT00106.1 KK852869 KDR14663.1 AJWK01005671 AE014297 ACZ95007.1 AF240470 BT003245 AAS65213.2 AGB96312.1 CH479186 EDW39155.1 CH480815 EDW43560.1 CH477310 EAT43969.1 JXUM01023257 KQ560656 KXJ81409.1 JXUM01071934 KQ562693 KXJ75323.1 CH954182 EDV53794.1 CM000160 EDW98881.2 GEBQ01009168 JAT30809.1 GEBQ01025049 JAT14928.1 KQ976737 KYM75926.1 KQ435878 KOX70021.1 GL440027 EFN66375.1 GGFJ01002531 MBW51672.1 ADTU01015830 KQ979763 KYN19253.1 CVRI01000072 CRL07504.1 CH964251 EDW83105.2 GGFJ01002529 MBW51670.1 AXCN02001149 GGFJ01002530 MBW51671.1 ADMH02001345 ETN62941.1 KQ982194 KYQ59045.1 GL888393 EGI61719.1 KQ981208 KYN44896.1 GBYB01008283 JAG78050.1 KQ976870 KYN07730.1 GL448301 EFN84844.1 CP012526 ALC47954.1 CH933806 EDW13937.2 KRG00736.1 NNAY01001330 OXU24334.1 GGFM01006077 MBW26828.1 CH940650 EDW66497.2 KRF82724.1 LJIJ01001406 ODM91835.1 GBXI01005937 JAD08355.1 APCN01000910 ACPB03024513 GGFK01003722 MBW37043.1 AAZX01002902 GGFK01003487 MBW36808.1 GGFK01003505 MBW36826.1 UFQS01000609 UFQT01000609 SSX05329.1 SSX25690.1 AXCM01000028 KZ288220 PBC32167.1 AAAB01008987 EAA01208.4 ATLV01018748 KE525248 KFB43337.1 GAMC01008291 JAB98264.1 GAMC01008290 JAB98265.1 GEDC01001996 JAS35302.1 GDHF01023615 JAI28699.1 GDHF01017436 JAI34878.1 GGFM01002272 MBW23023.1 GDHF01020329 JAI31985.1
NWSH01000740 PCG74475.1 RSAL01000315 RVE42617.1 AGBW02008599 OWR52935.1 KQ459602 KPI93310.1 KQ460615 KPJ13554.1 GDQN01006669 JAT84385.1 CH902617 EDV42482.1 GFDL01009692 JAV25353.1 OUUW01000005 SPP80154.1 GFDF01006664 JAV07420.1 GEZM01062164 JAV69994.1 GANO01000942 JAB58929.1 DS231831 EDS31216.1 KQ414706 KOC63093.1 KQ434889 KZC10347.1 KQ971345 EFA05049.2 CM000364 EDX14294.1 CM000070 EAL26678.4 GECU01007601 JAT00106.1 KK852869 KDR14663.1 AJWK01005671 AE014297 ACZ95007.1 AF240470 BT003245 AAS65213.2 AGB96312.1 CH479186 EDW39155.1 CH480815 EDW43560.1 CH477310 EAT43969.1 JXUM01023257 KQ560656 KXJ81409.1 JXUM01071934 KQ562693 KXJ75323.1 CH954182 EDV53794.1 CM000160 EDW98881.2 GEBQ01009168 JAT30809.1 GEBQ01025049 JAT14928.1 KQ976737 KYM75926.1 KQ435878 KOX70021.1 GL440027 EFN66375.1 GGFJ01002531 MBW51672.1 ADTU01015830 KQ979763 KYN19253.1 CVRI01000072 CRL07504.1 CH964251 EDW83105.2 GGFJ01002529 MBW51670.1 AXCN02001149 GGFJ01002530 MBW51671.1 ADMH02001345 ETN62941.1 KQ982194 KYQ59045.1 GL888393 EGI61719.1 KQ981208 KYN44896.1 GBYB01008283 JAG78050.1 KQ976870 KYN07730.1 GL448301 EFN84844.1 CP012526 ALC47954.1 CH933806 EDW13937.2 KRG00736.1 NNAY01001330 OXU24334.1 GGFM01006077 MBW26828.1 CH940650 EDW66497.2 KRF82724.1 LJIJ01001406 ODM91835.1 GBXI01005937 JAD08355.1 APCN01000910 ACPB03024513 GGFK01003722 MBW37043.1 AAZX01002902 GGFK01003487 MBW36808.1 GGFK01003505 MBW36826.1 UFQS01000609 UFQT01000609 SSX05329.1 SSX25690.1 AXCM01000028 KZ288220 PBC32167.1 AAAB01008987 EAA01208.4 ATLV01018748 KE525248 KFB43337.1 GAMC01008291 JAB98264.1 GAMC01008290 JAB98265.1 GEDC01001996 JAS35302.1 GDHF01023615 JAI28699.1 GDHF01017436 JAI34878.1 GGFM01002272 MBW23023.1 GDHF01020329 JAI31985.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000053240
+ More
UP000007801 UP000268350 UP000002320 UP000053825 UP000076502 UP000007266 UP000000304 UP000001819 UP000027135 UP000092461 UP000000803 UP000008744 UP000001292 UP000008820 UP000192221 UP000069940 UP000249989 UP000008711 UP000002282 UP000078540 UP000053105 UP000000311 UP000005205 UP000075901 UP000078492 UP000183832 UP000007798 UP000075886 UP000000673 UP000075809 UP000007755 UP000075880 UP000075884 UP000075885 UP000078541 UP000076408 UP000078542 UP000075902 UP000008237 UP000069272 UP000075881 UP000075920 UP000092553 UP000009192 UP000215335 UP000008792 UP000094527 UP000076407 UP000075840 UP000015103 UP000075903 UP000002358 UP000005203 UP000075900 UP000075883 UP000242457 UP000007062 UP000030765 UP000075882
UP000007801 UP000268350 UP000002320 UP000053825 UP000076502 UP000007266 UP000000304 UP000001819 UP000027135 UP000092461 UP000000803 UP000008744 UP000001292 UP000008820 UP000192221 UP000069940 UP000249989 UP000008711 UP000002282 UP000078540 UP000053105 UP000000311 UP000005205 UP000075901 UP000078492 UP000183832 UP000007798 UP000075886 UP000000673 UP000075809 UP000007755 UP000075880 UP000075884 UP000075885 UP000078541 UP000076408 UP000078542 UP000075902 UP000008237 UP000069272 UP000075881 UP000075920 UP000092553 UP000009192 UP000215335 UP000008792 UP000094527 UP000076407 UP000075840 UP000015103 UP000075903 UP000002358 UP000005203 UP000075900 UP000075883 UP000242457 UP000007062 UP000030765 UP000075882
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
H9JDY0
A0A2H1WCH3
S4PFY1
A0A2A4JR01
A0A437AWS1
A0A212FGR9
+ More
A0A194PQA0 A0A194R7R5 A0A1E1WBR6 B3LV10 A0A1Q3FCX7 A0A3B0JCQ0 A0A1L8DM59 A0A1Y1L8T5 U5EXI1 B0W373 A0A0L7QWU1 A0A154PEW5 D2A5N1 B4QTU0 Q29CG8 A0A1B6JLP8 A0A067QWF2 A0A1B0GHV0 E1JIV8 Q9VC27 Q7KS07 A0A0B4KHG2 B4GN29 B4HHL8 Q17CA6 A0A1W4VR20 A0A1W4W349 A0A1S4F865 A0A182H1V0 A0A182GLG6 B3P6X4 B4PV32 A0A1B6M4G2 A0A1B6KU33 A0A195AUS2 A0A0N0U3I5 E2AJI0 A0A2M4BF61 A0A158NHP2 A0A182SDC3 A0A195E285 A0A1J1J526 B4NFN8 A0A2M4BF39 A0A1Y9HAG1 A0A2M4BFD8 W5JFV8 A0A151XFD1 F4WVP9 A0A182JJR2 A0A1Y9H2V6 A0A240PL32 A0A195FWU1 A0A0C9Q2S0 A0A182Y3U6 A0A195D4A0 A0A182U8L9 E2BHJ1 A0A1I8JSS5 A0A240PKR8 A0A1Y9IVQ9 A0A0M4EHB4 B4K4I5 A0A0Q9WXI5 A0A232F0X7 A0A2M3ZEC1 B4LVZ2 A0A0Q9WP97 A0A1D2MFT3 A0A0A1XBN2 A0A1I8JVR0 A0A1I8JTD9 T1HZX0 A0A182UP50 A0A2M4A8B1 K7ISR9 A0A2M4A852 A0A2M4A7P4 A0A336KLF8 A0A088AD09 A0A1I8JUS5 A0A182MPD3 A0A2A3EKB5 Q7PY38 A0A1S4GCU3 A0A084VZE3 W8C6X3 W8BAG5 A0A1B6EBK1 A0A0K8UPS5 A0A0K8V810 A0A2M3Z3F3 A0A0K8V067 A0A182LA47
A0A194PQA0 A0A194R7R5 A0A1E1WBR6 B3LV10 A0A1Q3FCX7 A0A3B0JCQ0 A0A1L8DM59 A0A1Y1L8T5 U5EXI1 B0W373 A0A0L7QWU1 A0A154PEW5 D2A5N1 B4QTU0 Q29CG8 A0A1B6JLP8 A0A067QWF2 A0A1B0GHV0 E1JIV8 Q9VC27 Q7KS07 A0A0B4KHG2 B4GN29 B4HHL8 Q17CA6 A0A1W4VR20 A0A1W4W349 A0A1S4F865 A0A182H1V0 A0A182GLG6 B3P6X4 B4PV32 A0A1B6M4G2 A0A1B6KU33 A0A195AUS2 A0A0N0U3I5 E2AJI0 A0A2M4BF61 A0A158NHP2 A0A182SDC3 A0A195E285 A0A1J1J526 B4NFN8 A0A2M4BF39 A0A1Y9HAG1 A0A2M4BFD8 W5JFV8 A0A151XFD1 F4WVP9 A0A182JJR2 A0A1Y9H2V6 A0A240PL32 A0A195FWU1 A0A0C9Q2S0 A0A182Y3U6 A0A195D4A0 A0A182U8L9 E2BHJ1 A0A1I8JSS5 A0A240PKR8 A0A1Y9IVQ9 A0A0M4EHB4 B4K4I5 A0A0Q9WXI5 A0A232F0X7 A0A2M3ZEC1 B4LVZ2 A0A0Q9WP97 A0A1D2MFT3 A0A0A1XBN2 A0A1I8JVR0 A0A1I8JTD9 T1HZX0 A0A182UP50 A0A2M4A8B1 K7ISR9 A0A2M4A852 A0A2M4A7P4 A0A336KLF8 A0A088AD09 A0A1I8JUS5 A0A182MPD3 A0A2A3EKB5 Q7PY38 A0A1S4GCU3 A0A084VZE3 W8C6X3 W8BAG5 A0A1B6EBK1 A0A0K8UPS5 A0A0K8V810 A0A2M3Z3F3 A0A0K8V067 A0A182LA47
PDB
6IYC
E-value=3.68993e-47,
Score=477
Ontologies
GO
Topology
Subcellular location
Membrane
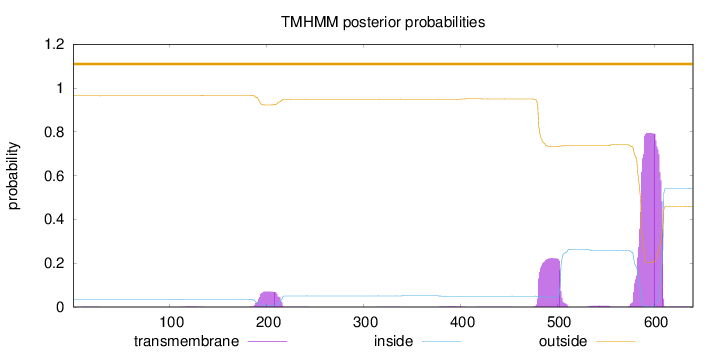
Length:
640
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
25.13814
Exp number, first 60 AAs:
0.00728
Total prob of N-in:
0.03565
outside
1 - 640
Population Genetic Test Statistics
Pi
283.940212
Theta
216.891823
Tajima's D
1.091345
CLR
0.067083
CSRT
0.679866006699665
Interpretation
Uncertain
