Pre Gene Modal
BGIBMGA007800
Annotation
PREDICTED:_uncharacterized_protein_LOC106712294_[Papilio_machaon]
Full name
E3 ubiquitin-protein ligase
Location in the cell
Extracellular Reliability : 1.801 Nuclear Reliability : 2.438
Sequence
CDS
ATGTCGGAGATAGTTGAAAATAAGGTTGACGAATTGCCAGACTTGGATAACTTACTTCAGTGTCCAGTATGTTATGAAATCCCTACAGGTCATATTTTCCAATGCAATGAAGGTCATAATGTATGTGGCAGATGTAAAGTTCGGCTTTCTGTCTGCCCGGTATGCAGGGCACTATTTTTTGGAACCAGGAACTATGCCATGGAAGAGCTCATTGCAAATGTCCGGAAACTAAGAGCTTTTAAATTAGGTGGAAAAGTTACAAGAGGATTATTAGCTTCAGATAGTAGTAGTACACCAGCAAGAGACAGTGGATCTGACAATGCAGAATATTTGGGTGAAGATGAAAATAACGATAATCAAAATTCAGTAGTGGCCCCTAACCCATGTAAAGGACTTTTTCGTTGCCTCTGCTGCAAGAATGGGAATTCCATCAGACTTCCAGCAGCTCGTTTACTTAACCACCTTCGCTACTTTCATTCTCCAGAACTTTTAGAGGTAAGTCTTTTCCCTAATCATTAG
Protein
MSEIVENKVDELPDLDNLLQCPVCYEIPTGHIFQCNEGHNVCGRCKVRLSVCPVCRALFFGTRNYAMEELIANVRKLRAFKLGGKVTRGLLASDSSSTPARDSGSDNAEYLGEDENNDNQNSVVAPNPCKGLFRCLCCKNGNSIRLPAARLLNHLRYFHSPELLEVSLFPNH
Summary
Description
E3 ubiquitin-protein ligase that mediates ubiquitination and subsequent proteasomal degradation of target proteins. E3 ubiquitin ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates.
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Similarity
Belongs to the SINA (Seven in absentia) family.
Uniprot
EC Number
2.3.2.27
Pubmed
EMBL
RSAL01000315
RVE42618.1
GDQN01001498
JAT89556.1
KQ460615
KPJ13553.1
+ More
KQ459602 KPI93309.1 AGBW02008599 OWR52936.1 NWSH01000740 PCG74476.1 ODYU01007669 SOQ50655.1 GAIX01012099 JAA80461.1 KQ979568 KYN21002.1 LBMM01005283 KMQ91655.1 ADTU01014960 ADTU01014961 KQ976580 KYM79955.1 GL888341 EGI62463.1 KQ982335 KYQ57659.1 GEZM01013542 JAV92494.1 KQ977721 KYN00365.1 GEBQ01024263 JAT15714.1 NNAY01000751 OXU26691.1 GDAI01000485 JAI17118.1 KK108395 QOIP01000006 EZA46720.1 RLU21253.1 GL441764 EFN64322.1 GECU01035638 GECU01009453 JAS72068.1 JAS98253.1 NNAY01001458 OXU23903.1
KQ459602 KPI93309.1 AGBW02008599 OWR52936.1 NWSH01000740 PCG74476.1 ODYU01007669 SOQ50655.1 GAIX01012099 JAA80461.1 KQ979568 KYN21002.1 LBMM01005283 KMQ91655.1 ADTU01014960 ADTU01014961 KQ976580 KYM79955.1 GL888341 EGI62463.1 KQ982335 KYQ57659.1 GEZM01013542 JAV92494.1 KQ977721 KYN00365.1 GEBQ01024263 JAT15714.1 NNAY01000751 OXU26691.1 GDAI01000485 JAI17118.1 KK108395 QOIP01000006 EZA46720.1 RLU21253.1 GL441764 EFN64322.1 GECU01035638 GECU01009453 JAS72068.1 JAS98253.1 NNAY01001458 OXU23903.1
Proteomes
Interpro
IPR013083
Znf_RING/FYVE/PHD
+ More
IPR001841 Znf_RING
IPR004162 SINA-like
IPR018121 7-in-absentia-prot_TRAF-dom
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR015919 Cadherin-like_sf
IPR017441 Protein_kinase_ATP_BS
IPR001841 Znf_RING
IPR004162 SINA-like
IPR018121 7-in-absentia-prot_TRAF-dom
IPR008266 Tyr_kinase_AS
IPR020635 Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR015919 Cadherin-like_sf
IPR017441 Protein_kinase_ATP_BS
Gene 3D
ProteinModelPortal
Ontologies
GO
PANTHER
Topology
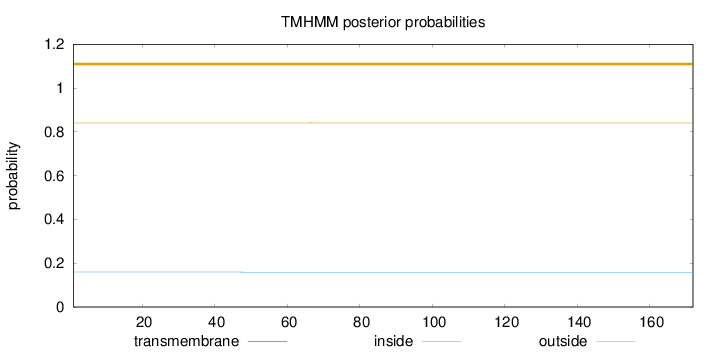
Length:
172
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00587999999999999
Exp number, first 60 AAs:
0.00359
Total prob of N-in:
0.15938
outside
1 - 172
Population Genetic Test Statistics
Pi
274.377517
Theta
203.022687
Tajima's D
1.254022
CLR
0
CSRT
0.723913804309784
Interpretation
Uncertain
