Pre Gene Modal
BGIBMGA007726
Annotation
PREDICTED:_mitochondrial_chaperone_BCS1_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 2.592
Sequence
CDS
ATGACTATCGTAGAATATATGGCTTCTTTAGCCCAAAATCCGTATTTTGGTGCAGGTTTTGGACTTTTCGGTGTAGGTGCAAGTGCAGCTATTCTACGAAAAGGATTGCAGACTTCAATGGTATTATTTAGAAGGCATTGTATGATAACCCTTGAAGTTCCATGCAGAGATAAATCCTATCAATGGCTATTACAATGGATTACACAAAAAGGAGCAAGAAAAACTCAACATCTAAGTGTCGAGACTTCTTTTGAACAAAAAGATTCTGGACAGATAAGAACTAAATACGACTTTATTCCTAGTGTTGGCCAACACTTTTTCCGATATGGTGGAACATGGATAAGAGTAGACCGTACAAGGGAACAGCAGACTTTGGACCTGCATATGGGGATACCATGGGAAACGGTAACATTAACTTCATTTGGCAGAAACAAGCAATTATATTATGGAATTCTAGAAGAAGCTAGAACAATGGCTCTCAAACAACATGAAGGTATGACAGTTATGTACACCGCAATGGGCTCTGAGTGGCGACCCTTTGGACACCCTCGTCGGCGCCGACCTATACACAGTGTAGTTCTTCGATCAGGATTAACAGAACGGATAGTTGCTGACTGTTTAGACTTCATTAATAACCCACAATGGTATACAGACCGTGGAATACCCTATAGAAGAGGCTATTTGCTTTATGGGCCACCGGGCTGCGGAAAATCTTCATTTATCATGGCATTAGCAGGAGAACTAGAATACAATATTTGTGTGCTCAACTTATCTGAACGAGGCTTAACAGATGATCGATTAAATCATTTGCTGAGTGTAGCTCCGCAACAGTCTATAATCTTACTTGAAGACATAGATGCGGCGTTTGTGTCCCGCGAGGACACGCCGACCCAGAAAGCGGCATACGAAGGTCTCAATCGCGTTACGTTTAGCGGACTGCTGAACTGTTTGGATGGCGTTGCGTCAACCGAAGCACGGATCGTCTTTATGACGACGAACTATTTGGAGAGGCTTGATCCGGCCCTGATCAGGCCAGGGCGTGTAGATATGAAAGAATACGTTGGATACTGCAACCAAGAACAGGTCGAACTAATGTTCTTAAGATTTTACAAAGGAGAAAAAGCGGCTGACCATGCTAAAACGTTTGCCGAAAAAGTAATGGAAGTAAAAAAAGAAGTCAGTCCAGCACAAATTCAAGGATACTTTATGTTTCATAAACATTCACCCCCAGAGGACGTATTGATGGATGTTAAATCAATATGGACGCTAGGATAA
Protein
MTIVEYMASLAQNPYFGAGFGLFGVGASAAILRKGLQTSMVLFRRHCMITLEVPCRDKSYQWLLQWITQKGARKTQHLSVETSFEQKDSGQIRTKYDFIPSVGQHFFRYGGTWIRVDRTREQQTLDLHMGIPWETVTLTSFGRNKQLYYGILEEARTMALKQHEGMTVMYTAMGSEWRPFGHPRRRRPIHSVVLRSGLTERIVADCLDFINNPQWYTDRGIPYRRGYLLYGPPGCGKSSFIMALAGELEYNICVLNLSERGLTDDRLNHLLSVAPQQSIILLEDIDAAFVSREDTPTQKAAYEGLNRVTFSGLLNCLDGVASTEARIVFMTTNYLERLDPALIRPGRVDMKEYVGYCNQEQVELMFLRFYKGEKAADHAKTFAEKVMEVKKEVSPAQIQGYFMFHKHSPPEDVLMDVKSIWTLG
Summary
Similarity
Belongs to the AAA ATPase family.
Uniprot
H9JDX9
A0A2A4JSW3
A0A3S2N9Z6
A0A2H1WCT3
A0A212FGS6
A0A194RCB6
+ More
A0A194PJ09 A0A2J7RJ42 D6WNU0 A0A067QYB5 N6UMK5 A0A1Y1M600 A0A182VI96 A0A182TXT2 Q7QAZ7 A0A1B6D208 Q16QY9 A0A182T8Y7 A0A182H7G7 A0A1W4XDY9 A0A182JFS7 A0A182PS69 A0A182PZC9 A0A182XX23 A0A182W176 A0A182LUU1 A0A084WP04 A0A182RX69 A0A182NCX3 W5J841 A0A1Q3F2T5 B0X1Q3 A0A158NNV1 A0A195BRZ4 A0A2A3EA90 A0A088AHF8 A0A151WJT2 A0A151IWT2 A0A026WQQ3 E2BYY8 A0A195EWC9 F4WVA8 A0A182FNI0 A0A151ICB0 A0A1B0D4U5 A0A182XG16 A0A182L6Z6 A0A0J7KU73 A0A182ID62 A0A0L7RJ00 A0A0L0BLC3 A0A1B6G2U4 A0A1I8PJS9 W8CA35 A0A1I8N285 A0A0M4E1G5 B4JPP2 A0A034W668 A0A0K8UKA4 B4KJK4 A0A1B6L7V4 B4MG54 B3MPN0 T1GR13 A0A1W4UUM5 B3N940 B4NZ64 A0A0R1DTJ6 A0A0N0BBP1 A0A1B0FRD3 D3TN96 B4HWH6 B4Q8Q9 Q9VL22 Q1EHJ2 A0A0A9XML0 A0A182JUK7 Q29NI5 A0A1A9UCR5 B4G799 A0A1A9ZX78 A0A154P694 A0A1A9WEW6 A0A1A9YQV4 A0A0Q9WW65 A0A1B0C1X0 E0VEP7 A0A023F3S6 T1HNS9 A0A3B0K6F3 A0A1J1IHZ8 A0A1B6I5J9 A0A0V0GC78 A0A069DT98 T1IN92 K7IM16 A0A1Z5KVV1 A0A310S4N5 A0A1S4HA67 A0A293LE41
A0A194PJ09 A0A2J7RJ42 D6WNU0 A0A067QYB5 N6UMK5 A0A1Y1M600 A0A182VI96 A0A182TXT2 Q7QAZ7 A0A1B6D208 Q16QY9 A0A182T8Y7 A0A182H7G7 A0A1W4XDY9 A0A182JFS7 A0A182PS69 A0A182PZC9 A0A182XX23 A0A182W176 A0A182LUU1 A0A084WP04 A0A182RX69 A0A182NCX3 W5J841 A0A1Q3F2T5 B0X1Q3 A0A158NNV1 A0A195BRZ4 A0A2A3EA90 A0A088AHF8 A0A151WJT2 A0A151IWT2 A0A026WQQ3 E2BYY8 A0A195EWC9 F4WVA8 A0A182FNI0 A0A151ICB0 A0A1B0D4U5 A0A182XG16 A0A182L6Z6 A0A0J7KU73 A0A182ID62 A0A0L7RJ00 A0A0L0BLC3 A0A1B6G2U4 A0A1I8PJS9 W8CA35 A0A1I8N285 A0A0M4E1G5 B4JPP2 A0A034W668 A0A0K8UKA4 B4KJK4 A0A1B6L7V4 B4MG54 B3MPN0 T1GR13 A0A1W4UUM5 B3N940 B4NZ64 A0A0R1DTJ6 A0A0N0BBP1 A0A1B0FRD3 D3TN96 B4HWH6 B4Q8Q9 Q9VL22 Q1EHJ2 A0A0A9XML0 A0A182JUK7 Q29NI5 A0A1A9UCR5 B4G799 A0A1A9ZX78 A0A154P694 A0A1A9WEW6 A0A1A9YQV4 A0A0Q9WW65 A0A1B0C1X0 E0VEP7 A0A023F3S6 T1HNS9 A0A3B0K6F3 A0A1J1IHZ8 A0A1B6I5J9 A0A0V0GC78 A0A069DT98 T1IN92 K7IM16 A0A1Z5KVV1 A0A310S4N5 A0A1S4HA67 A0A293LE41
Pubmed
19121390
22118469
26354079
18362917
19820115
24845553
+ More
23537049 28004739 12364791 14747013 17210077 17510324 26483478 25244985 24438588 20920257 23761445 21347285 24508170 30249741 20798317 21719571 20966253 26108605 24495485 25315136 17994087 25348373 18057021 17550304 20353571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26823975 15632085 20566863 25474469 26334808 20075255 28528879 9087549
23537049 28004739 12364791 14747013 17210077 17510324 26483478 25244985 24438588 20920257 23761445 21347285 24508170 30249741 20798317 21719571 20966253 26108605 24495485 25315136 17994087 25348373 18057021 17550304 20353571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25401762 26823975 15632085 20566863 25474469 26334808 20075255 28528879 9087549
EMBL
BABH01002603
NWSH01000740
PCG74480.1
RSAL01000370
RVE42146.1
ODYU01007669
+ More
SOQ50652.1 AGBW02008599 OWR52938.1 KQ460615 KPJ13551.1 KQ459602 KPI93307.1 NEVH01003007 PNF40850.1 KQ971342 EFA03729.1 KK852880 KDR14433.1 APGK01026292 KB740624 KB632335 ENN79957.1 ERL92797.1 GEZM01039704 JAV81279.1 AAAB01008880 EAA08672.3 GEDC01017655 JAS19643.1 CH477727 EAT36833.1 JXUM01116198 KQ565958 KXJ70494.1 AXCN02001559 AXCM01004697 ATLV01024805 KE525361 KFB51948.1 ADMH02002109 ETN58949.1 GFDL01013170 JAV21875.1 DS232266 EDS38781.1 ADTU01021804 KQ976417 KYM89785.1 KZ288310 PBC28647.1 KQ983031 KYQ48118.1 KQ980843 KYN12289.1 KK107128 QOIP01000006 EZA58377.1 RLU21279.1 GL451531 EFN79106.1 KQ981953 KYN32456.1 GL888384 EGI61874.1 KQ978062 KYM97716.1 AJVK01024878 LBMM01003210 KMQ93804.1 APCN01005034 KQ414583 KOC70783.1 JRES01001695 KNC20885.1 GECZ01013025 JAS56744.1 GAMC01001619 JAC04937.1 CP012523 ALC39204.1 CH916372 EDV98872.1 GAKP01009694 JAC49258.1 GDHF01025569 JAI26745.1 CH933807 EDW13584.1 GEBQ01020185 JAT19792.1 CH940671 EDW58192.1 KRF78172.1 KRF78173.1 CH902620 EDV32278.2 KPU73876.1 KPU73877.1 CAQQ02395036 CH954177 EDV58475.1 KQS70410.1 CM000157 EDW88759.1 KRJ98004.1 KRJ98005.1 KRJ98006.1 KQ435969 KOX67854.1 CCAG010018514 EZ422898 ADD19174.1 CH480818 EDW52371.1 CM000361 CM002910 EDX04474.1 KMY89430.1 AE014134 AY089691 AAF52876.1 AAL90429.1 AY075442 ABF85762.1 GBHO01021645 GBHO01021643 GBHO01021642 GBRD01016721 GBRD01016719 GBRD01009209 GDHC01019892 GDHC01005207 JAG21959.1 JAG21961.1 JAG21962.1 JAG49106.1 JAP98736.1 JAQ13422.1 CH379059 EAL34235.2 CH479180 EDW28353.1 KQ434826 KZC07449.1 CH964282 KRG00236.1 JXJN01020904 JXJN01024195 DS235093 EEB11853.1 GBBI01002666 JAC16046.1 ACPB03018091 OUUW01000006 SPP81589.1 CVRI01000053 CRK99824.1 GECU01025502 JAS82204.1 GECL01000401 JAP05723.1 GBGD01001714 JAC87175.1 JH431152 GFJQ02007862 JAV99107.1 KQ775960 OAD52232.1 AAAB01001641 GFWV01002263 MAA26993.1
SOQ50652.1 AGBW02008599 OWR52938.1 KQ460615 KPJ13551.1 KQ459602 KPI93307.1 NEVH01003007 PNF40850.1 KQ971342 EFA03729.1 KK852880 KDR14433.1 APGK01026292 KB740624 KB632335 ENN79957.1 ERL92797.1 GEZM01039704 JAV81279.1 AAAB01008880 EAA08672.3 GEDC01017655 JAS19643.1 CH477727 EAT36833.1 JXUM01116198 KQ565958 KXJ70494.1 AXCN02001559 AXCM01004697 ATLV01024805 KE525361 KFB51948.1 ADMH02002109 ETN58949.1 GFDL01013170 JAV21875.1 DS232266 EDS38781.1 ADTU01021804 KQ976417 KYM89785.1 KZ288310 PBC28647.1 KQ983031 KYQ48118.1 KQ980843 KYN12289.1 KK107128 QOIP01000006 EZA58377.1 RLU21279.1 GL451531 EFN79106.1 KQ981953 KYN32456.1 GL888384 EGI61874.1 KQ978062 KYM97716.1 AJVK01024878 LBMM01003210 KMQ93804.1 APCN01005034 KQ414583 KOC70783.1 JRES01001695 KNC20885.1 GECZ01013025 JAS56744.1 GAMC01001619 JAC04937.1 CP012523 ALC39204.1 CH916372 EDV98872.1 GAKP01009694 JAC49258.1 GDHF01025569 JAI26745.1 CH933807 EDW13584.1 GEBQ01020185 JAT19792.1 CH940671 EDW58192.1 KRF78172.1 KRF78173.1 CH902620 EDV32278.2 KPU73876.1 KPU73877.1 CAQQ02395036 CH954177 EDV58475.1 KQS70410.1 CM000157 EDW88759.1 KRJ98004.1 KRJ98005.1 KRJ98006.1 KQ435969 KOX67854.1 CCAG010018514 EZ422898 ADD19174.1 CH480818 EDW52371.1 CM000361 CM002910 EDX04474.1 KMY89430.1 AE014134 AY089691 AAF52876.1 AAL90429.1 AY075442 ABF85762.1 GBHO01021645 GBHO01021643 GBHO01021642 GBRD01016721 GBRD01016719 GBRD01009209 GDHC01019892 GDHC01005207 JAG21959.1 JAG21961.1 JAG21962.1 JAG49106.1 JAP98736.1 JAQ13422.1 CH379059 EAL34235.2 CH479180 EDW28353.1 KQ434826 KZC07449.1 CH964282 KRG00236.1 JXJN01020904 JXJN01024195 DS235093 EEB11853.1 GBBI01002666 JAC16046.1 ACPB03018091 OUUW01000006 SPP81589.1 CVRI01000053 CRK99824.1 GECU01025502 JAS82204.1 GECL01000401 JAP05723.1 GBGD01001714 JAC87175.1 JH431152 GFJQ02007862 JAV99107.1 KQ775960 OAD52232.1 AAAB01001641 GFWV01002263 MAA26993.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000235965 UP000007266 UP000027135 UP000019118 UP000030742 UP000075903 UP000075902 UP000007062 UP000008820 UP000075901 UP000069940 UP000249989 UP000192223 UP000075880 UP000075885 UP000075886 UP000076408 UP000075920 UP000075883 UP000030765 UP000075900 UP000075884 UP000000673 UP000002320 UP000005205 UP000078540 UP000242457 UP000005203 UP000075809 UP000078492 UP000053097 UP000279307 UP000008237 UP000078541 UP000007755 UP000069272 UP000078542 UP000092462 UP000076407 UP000075882 UP000036403 UP000075840 UP000053825 UP000037069 UP000095300 UP000095301 UP000092553 UP000001070 UP000009192 UP000008792 UP000007801 UP000015102 UP000192221 UP000008711 UP000002282 UP000053105 UP000092444 UP000001292 UP000000304 UP000000803 UP000075881 UP000001819 UP000078200 UP000008744 UP000092445 UP000076502 UP000091820 UP000092443 UP000007798 UP000092460 UP000009046 UP000015103 UP000268350 UP000183832 UP000002358
UP000235965 UP000007266 UP000027135 UP000019118 UP000030742 UP000075903 UP000075902 UP000007062 UP000008820 UP000075901 UP000069940 UP000249989 UP000192223 UP000075880 UP000075885 UP000075886 UP000076408 UP000075920 UP000075883 UP000030765 UP000075900 UP000075884 UP000000673 UP000002320 UP000005205 UP000078540 UP000242457 UP000005203 UP000075809 UP000078492 UP000053097 UP000279307 UP000008237 UP000078541 UP000007755 UP000069272 UP000078542 UP000092462 UP000076407 UP000075882 UP000036403 UP000075840 UP000053825 UP000037069 UP000095300 UP000095301 UP000092553 UP000001070 UP000009192 UP000008792 UP000007801 UP000015102 UP000192221 UP000008711 UP000002282 UP000053105 UP000092444 UP000001292 UP000000304 UP000000803 UP000075881 UP000001819 UP000078200 UP000008744 UP000092445 UP000076502 UP000091820 UP000092443 UP000007798 UP000092460 UP000009046 UP000015103 UP000268350 UP000183832 UP000002358
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9JDX9
A0A2A4JSW3
A0A3S2N9Z6
A0A2H1WCT3
A0A212FGS6
A0A194RCB6
+ More
A0A194PJ09 A0A2J7RJ42 D6WNU0 A0A067QYB5 N6UMK5 A0A1Y1M600 A0A182VI96 A0A182TXT2 Q7QAZ7 A0A1B6D208 Q16QY9 A0A182T8Y7 A0A182H7G7 A0A1W4XDY9 A0A182JFS7 A0A182PS69 A0A182PZC9 A0A182XX23 A0A182W176 A0A182LUU1 A0A084WP04 A0A182RX69 A0A182NCX3 W5J841 A0A1Q3F2T5 B0X1Q3 A0A158NNV1 A0A195BRZ4 A0A2A3EA90 A0A088AHF8 A0A151WJT2 A0A151IWT2 A0A026WQQ3 E2BYY8 A0A195EWC9 F4WVA8 A0A182FNI0 A0A151ICB0 A0A1B0D4U5 A0A182XG16 A0A182L6Z6 A0A0J7KU73 A0A182ID62 A0A0L7RJ00 A0A0L0BLC3 A0A1B6G2U4 A0A1I8PJS9 W8CA35 A0A1I8N285 A0A0M4E1G5 B4JPP2 A0A034W668 A0A0K8UKA4 B4KJK4 A0A1B6L7V4 B4MG54 B3MPN0 T1GR13 A0A1W4UUM5 B3N940 B4NZ64 A0A0R1DTJ6 A0A0N0BBP1 A0A1B0FRD3 D3TN96 B4HWH6 B4Q8Q9 Q9VL22 Q1EHJ2 A0A0A9XML0 A0A182JUK7 Q29NI5 A0A1A9UCR5 B4G799 A0A1A9ZX78 A0A154P694 A0A1A9WEW6 A0A1A9YQV4 A0A0Q9WW65 A0A1B0C1X0 E0VEP7 A0A023F3S6 T1HNS9 A0A3B0K6F3 A0A1J1IHZ8 A0A1B6I5J9 A0A0V0GC78 A0A069DT98 T1IN92 K7IM16 A0A1Z5KVV1 A0A310S4N5 A0A1S4HA67 A0A293LE41
A0A194PJ09 A0A2J7RJ42 D6WNU0 A0A067QYB5 N6UMK5 A0A1Y1M600 A0A182VI96 A0A182TXT2 Q7QAZ7 A0A1B6D208 Q16QY9 A0A182T8Y7 A0A182H7G7 A0A1W4XDY9 A0A182JFS7 A0A182PS69 A0A182PZC9 A0A182XX23 A0A182W176 A0A182LUU1 A0A084WP04 A0A182RX69 A0A182NCX3 W5J841 A0A1Q3F2T5 B0X1Q3 A0A158NNV1 A0A195BRZ4 A0A2A3EA90 A0A088AHF8 A0A151WJT2 A0A151IWT2 A0A026WQQ3 E2BYY8 A0A195EWC9 F4WVA8 A0A182FNI0 A0A151ICB0 A0A1B0D4U5 A0A182XG16 A0A182L6Z6 A0A0J7KU73 A0A182ID62 A0A0L7RJ00 A0A0L0BLC3 A0A1B6G2U4 A0A1I8PJS9 W8CA35 A0A1I8N285 A0A0M4E1G5 B4JPP2 A0A034W668 A0A0K8UKA4 B4KJK4 A0A1B6L7V4 B4MG54 B3MPN0 T1GR13 A0A1W4UUM5 B3N940 B4NZ64 A0A0R1DTJ6 A0A0N0BBP1 A0A1B0FRD3 D3TN96 B4HWH6 B4Q8Q9 Q9VL22 Q1EHJ2 A0A0A9XML0 A0A182JUK7 Q29NI5 A0A1A9UCR5 B4G799 A0A1A9ZX78 A0A154P694 A0A1A9WEW6 A0A1A9YQV4 A0A0Q9WW65 A0A1B0C1X0 E0VEP7 A0A023F3S6 T1HNS9 A0A3B0K6F3 A0A1J1IHZ8 A0A1B6I5J9 A0A0V0GC78 A0A069DT98 T1IN92 K7IM16 A0A1Z5KVV1 A0A310S4N5 A0A1S4HA67 A0A293LE41
PDB
5VHS
E-value=1.87138e-14,
Score=193
Ontologies
GO
PANTHER
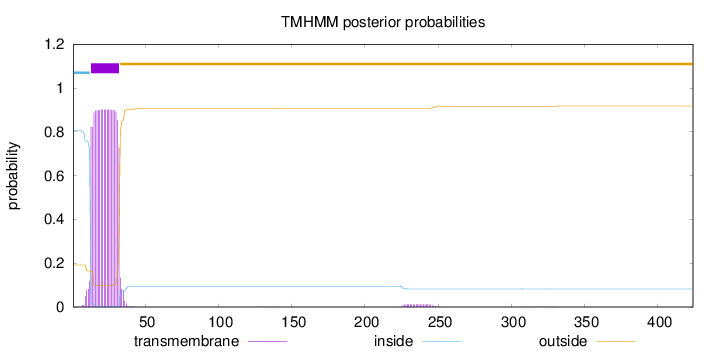
Topology
Length:
424
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.57972
Exp number, first 60 AAs:
18.30959
Total prob of N-in:
0.80790
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 32
outside
33 - 424
Population Genetic Test Statistics
Pi
269.428462
Theta
192.108807
Tajima's D
1.278545
CLR
0
CSRT
0.731613419329034
Interpretation
Uncertain
