Gene
KWMTBOMO08946
Annotation
PREDICTED:_uncharacterized_protein_LOC106138667_[Amyelois_transitella]
Full name
Transcription termination factor 5, mitochondrial
Alternative Name
Mitochondrial transcription termination factor 5
Location in the cell
Nuclear Reliability : 3.388
Sequence
CDS
ATGGATAATTATGGAGAAATTTTAAGAGAATGTCAATTCATCAAAATTATAACCAAACACATTATCAAATATCACACTCTTGTAAGATCCAGGACAATCGCCAAATTAAAAAAGGATGGTTTGATCAAAGCAGATCTAAATCTAGAAAAGGTTTTACAAGACTGTTTTCATGACTGGCCAGAGAGTAATAAAAGATTGCATCATTTTACAGATTCAACCACATCTATTTTACAAGTTCGTATGTGCGTATTGGAAAACTATTTGAAATGGAGATTGTCCATTGAAACTGACGAGTTCCAAAAATACTGTAAAAATTACATGTCTTTCAAACATAAACCAATGTCAGACATTCGAGAAGCTTTAGATATTGCACAAAATGTTATCAAGTTCGATACTGAAGTTATCAGACGCAATGGGTTTATTATCTCATCTGATCCGGTTAATACTAAATTGATACTAGAAAATGTCGAATCGTTAGCTGGCTTTGAAATACAGGACGCTATAAAAATCGAACCTGCCATCTTAAAAAATAACTATAATGCTTTGTTAGAAATGAAGAGTATTTTAGAGGAATACAGAATACCATTGGAGGCTCAAAGACGCTGTTTAAAAATATATTGCATGTGCCCGGAAACAGTACGCGAGCGACTAAAAGAACTAGCCAGTTTAAAAGAATATCAAGTATTATCTACCAACCCTAGAGTATTATCAATGGTCGTTCATAAGAAAAAAATGTTATCACGATTAAGTAAGATACAGGCAGCGAAAAAACAATGTTACAGTCTCAATCATCTCATCTCGTCCAGTAAAGTATTTAACAATTACATAAGCAATTTCGGTAATAAAGTCTGTGGTAGAGATATCGCAATACTAATAAGTTCTTCATTGAATCGGGAGGGATTCGATCATAATAAAGAGATAAGCGAATCCGAGAGGTTGAAGGAGGTGTTACAGCAGTTGAGAAGGCACAAGTTCTGGCTGCACACCGCGTTGGACGTGGTCTGCGACAATTTGCAGTTTCTGAAAAGTCGATTCAACGACAGGGATATTATCAAAAACTGTCAGCTGGTTTTGTATCCTGTAACTGAAATAGAACAATATATAACTAAATTATTAGACATTCGAGACAATAAAGAAATCCATTCACAGGACCGAGGAAATATGTATTACAATAATTTGAATTATGAAAAGCTAACTGGTGAACAGATTCTGAGTTTAGTATTATATGAGATAGAGAAGAAGTATCATTTTAGCGGAGATGGAATATGGGGAATGCAGGACGGTGTTAAAGTGGAATCGCGGGTCTAA
Protein
MDNYGEILRECQFIKIITKHIIKYHTLVRSRTIAKLKKDGLIKADLNLEKVLQDCFHDWPESNKRLHHFTDSTTSILQVRMCVLENYLKWRLSIETDEFQKYCKNYMSFKHKPMSDIREALDIAQNVIKFDTEVIRRNGFIISSDPVNTKLILENVESLAGFEIQDAIKIEPAILKNNYNALLEMKSILEEYRIPLEAQRRCLKIYCMCPETVRERLKELASLKEYQVLSTNPRVLSMVVHKKKMLSRLSKIQAAKKQCYSLNHLISSSKVFNNYISNFGNKVCGRDIAILISSSLNREGFDHNKEISESERLKEVLQQLRRHKFWLHTALDVVCDNLQFLKSRFNDRDIIKNCQLVLYPVTEIEQYITKLLDIRDNKEIHSQDRGNMYYNNLNYEKLTGEQILSLVLYEIEKKYHFSGDGIWGMQDGVKVESRV
Summary
Description
Binds promoter DNA and regulates initiation of transcription (PubMed:22784680, PubMed:24068965). Regulates mitochondrial replication and transcription (PubMed:22784680, PubMed:24068965). Required for normal topology and maintenance of mitochondrial DNA (mtDNA) levels (PubMed:22784680, PubMed:24068965). Regulates mtDNA replication by re-activating replication after replication pausing (PubMed:24068965). Likely to regulate replication pausing by coordinating with the mitochondrial termination factor mTTF which promotes replication pausing (PubMed:24068965). Their function in replication pausing prevents unregulated replication that may occur for example by collisions between the machineries of DNA replication and transcription during mtDNA synthesis (PubMed:24068965). This ensures the incorporation of RNA transcripts into replication intermediates at the replication fork and allows for proper fork progression (PubMed:24068965). Possibly functions downstream of Dref which activates genes involved in mtDNA replication and maintenance (PubMed:24068965).
Subunit
Probably binds to the mTTF-DNA complex.
Similarity
Belongs to the mTERF family.
Keywords
Complete proteome
DNA replication
Mitochondrion
Reference proteome
Transcription
Transcription regulation
Transit peptide
Feature
chain Transcription termination factor 5, mitochondrial
Uniprot
A0A1E1W3M8
A0A2H1X0K1
A0A2A4JDR3
A0A194PJV2
A0A3S2LR69
A0A0L7LD78
+ More
H9JE35 A0A194R8L6 A0A0L7LVA7 A0A212F6P2 Q17F49 A0A1S4F521 B4NG99 A0A1I8Q2P6 A0A1I8N5F7 B4K7H0 A0A3B0JNY9 A0A182JJV1 B4GLN0 Q293T7 B4JI56 W5JR45 B3MTR5 B4M0W8 B4IB88 A0A182H7T3 Q9VEB4 A0A1W4U8U7 B4PL83 A0A1B0CTU3 A0A0L0BX74 A0A1L8D7Q0 B3NZ95 A0A182U5G8 A0A2C9H8J7 A0A182HZA8 Q7QAR2 A0A182MU92 A0A182KXI1 A0A182QNZ0 A0A182RZG6 A0A1B6CTF5 A0A182PS21 W8BXM5 A0A0A1WN33 A0A182VJJ5 A0A1A9UCL3 A0A1B0A628 A0A1A9X8P0 A0A1B0GEC2 A0A336MF22 A0A0K8VK14 A0A1B0BFD1 A0A182K3M2 A0A182Y7D8 A0A182VQ52 A0A1J1HUD3 A0A034WT76 A0A1Q3F2E9 A0A2J7Q1Z9 A0A2J7Q1Z1 B0XD92 A0A067R7T8 A0A2J7Q1Y6 Q17F48 A0A1S4F508 A0A1A9X216 A0A182NLH0 J9JXS7 A0A2S2QG36 A0A226E2A6 A0A1J1IF61 B4QUI0 A0A182FT12 A0A1D2N4P7 A0A293LR41 D6WKL7 A0A0A9WVV1 A0A0C9RB16 A0A1B6KIH5 A0A131YIM6 A0A224Z1C6 L7LUP4 T1HPB6 A0A026VU53 A0A131XAK6 A0A3L8E355
H9JE35 A0A194R8L6 A0A0L7LVA7 A0A212F6P2 Q17F49 A0A1S4F521 B4NG99 A0A1I8Q2P6 A0A1I8N5F7 B4K7H0 A0A3B0JNY9 A0A182JJV1 B4GLN0 Q293T7 B4JI56 W5JR45 B3MTR5 B4M0W8 B4IB88 A0A182H7T3 Q9VEB4 A0A1W4U8U7 B4PL83 A0A1B0CTU3 A0A0L0BX74 A0A1L8D7Q0 B3NZ95 A0A182U5G8 A0A2C9H8J7 A0A182HZA8 Q7QAR2 A0A182MU92 A0A182KXI1 A0A182QNZ0 A0A182RZG6 A0A1B6CTF5 A0A182PS21 W8BXM5 A0A0A1WN33 A0A182VJJ5 A0A1A9UCL3 A0A1B0A628 A0A1A9X8P0 A0A1B0GEC2 A0A336MF22 A0A0K8VK14 A0A1B0BFD1 A0A182K3M2 A0A182Y7D8 A0A182VQ52 A0A1J1HUD3 A0A034WT76 A0A1Q3F2E9 A0A2J7Q1Z9 A0A2J7Q1Z1 B0XD92 A0A067R7T8 A0A2J7Q1Y6 Q17F48 A0A1S4F508 A0A1A9X216 A0A182NLH0 J9JXS7 A0A2S2QG36 A0A226E2A6 A0A1J1IF61 B4QUI0 A0A182FT12 A0A1D2N4P7 A0A293LR41 D6WKL7 A0A0A9WVV1 A0A0C9RB16 A0A1B6KIH5 A0A131YIM6 A0A224Z1C6 L7LUP4 T1HPB6 A0A026VU53 A0A131XAK6 A0A3L8E355
Pubmed
26354079
26227816
19121390
22118469
17510324
17994087
+ More
25315136 15632085 20920257 23761445 26483478 10731132 12537572 12537569 22784680 24068965 17550304 26108605 12364791 14747013 17210077 20966253 24495485 25830018 25244985 25348373 24845553 27289101 18362917 19820115 25401762 26830274 28797301 25576852 24508170 28049606 30249741
25315136 15632085 20920257 23761445 26483478 10731132 12537572 12537569 22784680 24068965 17550304 26108605 12364791 14747013 17210077 20966253 24495485 25830018 25244985 25348373 24845553 27289101 18362917 19820115 25401762 26830274 28797301 25576852 24508170 28049606 30249741
EMBL
GDQN01009484
JAT81570.1
ODYU01012526
SOQ58890.1
NWSH01001761
PCG70225.1
+ More
KQ459602 KPI93303.1 RSAL01000370 RVE42143.1 JTDY01001580 KOB73468.1 BABH01002535 KQ460615 KPJ13590.1 JTDY01000017 KOB79397.1 AGBW02009983 OWR49405.1 CH477276 EAT45134.1 CH964251 EDW83316.1 CH933806 EDW15314.1 OUUW01000008 SPP83845.1 CH479185 EDW38454.1 CM000070 EAL29127.2 CH916369 EDV92937.1 ADMH02000418 ETN66596.1 CH902623 EDV30196.1 CH940650 EDW68427.1 CH480827 EDW44646.1 JXUM01116959 KQ566037 KXJ70443.1 AE014297 AY061101 AAL28649.1 CM000160 EDW95865.1 AJWK01027994 JRES01001197 KNC24658.1 GFDF01011591 JAV02493.1 CH954181 EDV48637.1 APCN01005522 AAAB01008888 EAA08774.4 AXCM01001356 AXCN02000136 GEDC01020548 JAS16750.1 GAMC01002498 JAC04058.1 GBXI01014504 JAC99787.1 CCAG010012182 UFQT01001076 SSX28766.1 GDHF01013404 JAI38910.1 JXJN01013371 CVRI01000020 CRK91162.1 GAKP01001103 JAC57849.1 GFDL01013324 JAV21721.1 NEVH01019374 PNF22606.1 PNF22604.1 DS232745 EDS45346.1 KK852880 KDR14429.1 PNF22605.1 EAT45135.1 ABLF02040270 GGMS01007512 MBY76715.1 LNIX01000007 OXA51559.1 CVRI01000047 CRK98188.1 CM000364 EDX12480.1 LJIJ01000247 ODM99915.1 GFWV01006254 MAA30984.1 KQ971342 EFA03014.2 GBHO01033006 GBRD01010143 JAG10598.1 JAG55681.1 GBYB01005510 JAG75277.1 GEBQ01028728 JAT11249.1 GEDV01009423 JAP79134.1 GFPF01008674 MAA19820.1 GACK01009707 JAA55327.1 ACPB03014848 KK107928 EZA47170.1 GEFH01004458 JAP64123.1 QOIP01000001 RLU27134.1
KQ459602 KPI93303.1 RSAL01000370 RVE42143.1 JTDY01001580 KOB73468.1 BABH01002535 KQ460615 KPJ13590.1 JTDY01000017 KOB79397.1 AGBW02009983 OWR49405.1 CH477276 EAT45134.1 CH964251 EDW83316.1 CH933806 EDW15314.1 OUUW01000008 SPP83845.1 CH479185 EDW38454.1 CM000070 EAL29127.2 CH916369 EDV92937.1 ADMH02000418 ETN66596.1 CH902623 EDV30196.1 CH940650 EDW68427.1 CH480827 EDW44646.1 JXUM01116959 KQ566037 KXJ70443.1 AE014297 AY061101 AAL28649.1 CM000160 EDW95865.1 AJWK01027994 JRES01001197 KNC24658.1 GFDF01011591 JAV02493.1 CH954181 EDV48637.1 APCN01005522 AAAB01008888 EAA08774.4 AXCM01001356 AXCN02000136 GEDC01020548 JAS16750.1 GAMC01002498 JAC04058.1 GBXI01014504 JAC99787.1 CCAG010012182 UFQT01001076 SSX28766.1 GDHF01013404 JAI38910.1 JXJN01013371 CVRI01000020 CRK91162.1 GAKP01001103 JAC57849.1 GFDL01013324 JAV21721.1 NEVH01019374 PNF22606.1 PNF22604.1 DS232745 EDS45346.1 KK852880 KDR14429.1 PNF22605.1 EAT45135.1 ABLF02040270 GGMS01007512 MBY76715.1 LNIX01000007 OXA51559.1 CVRI01000047 CRK98188.1 CM000364 EDX12480.1 LJIJ01000247 ODM99915.1 GFWV01006254 MAA30984.1 KQ971342 EFA03014.2 GBHO01033006 GBRD01010143 JAG10598.1 JAG55681.1 GBYB01005510 JAG75277.1 GEBQ01028728 JAT11249.1 GEDV01009423 JAP79134.1 GFPF01008674 MAA19820.1 GACK01009707 JAA55327.1 ACPB03014848 KK107928 EZA47170.1 GEFH01004458 JAP64123.1 QOIP01000001 RLU27134.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000037510
UP000005204
UP000053240
+ More
UP000007151 UP000008820 UP000007798 UP000095300 UP000095301 UP000009192 UP000268350 UP000075880 UP000008744 UP000001819 UP000001070 UP000000673 UP000007801 UP000008792 UP000001292 UP000069940 UP000249989 UP000000803 UP000192221 UP000002282 UP000092461 UP000037069 UP000008711 UP000075902 UP000076407 UP000075840 UP000007062 UP000075883 UP000075882 UP000075886 UP000075900 UP000075885 UP000075903 UP000078200 UP000092445 UP000092443 UP000092444 UP000092460 UP000075881 UP000076408 UP000075920 UP000183832 UP000235965 UP000002320 UP000027135 UP000091820 UP000075884 UP000007819 UP000198287 UP000000304 UP000069272 UP000094527 UP000007266 UP000015103 UP000053097 UP000279307
UP000007151 UP000008820 UP000007798 UP000095300 UP000095301 UP000009192 UP000268350 UP000075880 UP000008744 UP000001819 UP000001070 UP000000673 UP000007801 UP000008792 UP000001292 UP000069940 UP000249989 UP000000803 UP000192221 UP000002282 UP000092461 UP000037069 UP000008711 UP000075902 UP000076407 UP000075840 UP000007062 UP000075883 UP000075882 UP000075886 UP000075900 UP000075885 UP000075903 UP000078200 UP000092445 UP000092443 UP000092444 UP000092460 UP000075881 UP000076408 UP000075920 UP000183832 UP000235965 UP000002320 UP000027135 UP000091820 UP000075884 UP000007819 UP000198287 UP000000304 UP000069272 UP000094527 UP000007266 UP000015103 UP000053097 UP000279307
Interpro
IPR003690
MTERF
+ More
IPR038538 MTERF_sf
IPR001494 Importin-beta_N
IPR016024 ARM-type_fold
IPR013598 Exportin-1/Importin-b-like
IPR011989 ARM-like
IPR036928 AS_sf
IPR020556 Amidase_CS
IPR023631 Amidase_dom
IPR000717 PCI_dom
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR026736 Virilizer
IPR031801 VIR_N
IPR036187 DNA_mismatch_repair_MutS_sf
IPR038538 MTERF_sf
IPR001494 Importin-beta_N
IPR016024 ARM-type_fold
IPR013598 Exportin-1/Importin-b-like
IPR011989 ARM-like
IPR036928 AS_sf
IPR020556 Amidase_CS
IPR023631 Amidase_dom
IPR000717 PCI_dom
IPR036390 WH_DNA-bd_sf
IPR036388 WH-like_DNA-bd_sf
IPR026736 Virilizer
IPR031801 VIR_N
IPR036187 DNA_mismatch_repair_MutS_sf
Gene 3D
ProteinModelPortal
A0A1E1W3M8
A0A2H1X0K1
A0A2A4JDR3
A0A194PJV2
A0A3S2LR69
A0A0L7LD78
+ More
H9JE35 A0A194R8L6 A0A0L7LVA7 A0A212F6P2 Q17F49 A0A1S4F521 B4NG99 A0A1I8Q2P6 A0A1I8N5F7 B4K7H0 A0A3B0JNY9 A0A182JJV1 B4GLN0 Q293T7 B4JI56 W5JR45 B3MTR5 B4M0W8 B4IB88 A0A182H7T3 Q9VEB4 A0A1W4U8U7 B4PL83 A0A1B0CTU3 A0A0L0BX74 A0A1L8D7Q0 B3NZ95 A0A182U5G8 A0A2C9H8J7 A0A182HZA8 Q7QAR2 A0A182MU92 A0A182KXI1 A0A182QNZ0 A0A182RZG6 A0A1B6CTF5 A0A182PS21 W8BXM5 A0A0A1WN33 A0A182VJJ5 A0A1A9UCL3 A0A1B0A628 A0A1A9X8P0 A0A1B0GEC2 A0A336MF22 A0A0K8VK14 A0A1B0BFD1 A0A182K3M2 A0A182Y7D8 A0A182VQ52 A0A1J1HUD3 A0A034WT76 A0A1Q3F2E9 A0A2J7Q1Z9 A0A2J7Q1Z1 B0XD92 A0A067R7T8 A0A2J7Q1Y6 Q17F48 A0A1S4F508 A0A1A9X216 A0A182NLH0 J9JXS7 A0A2S2QG36 A0A226E2A6 A0A1J1IF61 B4QUI0 A0A182FT12 A0A1D2N4P7 A0A293LR41 D6WKL7 A0A0A9WVV1 A0A0C9RB16 A0A1B6KIH5 A0A131YIM6 A0A224Z1C6 L7LUP4 T1HPB6 A0A026VU53 A0A131XAK6 A0A3L8E355
H9JE35 A0A194R8L6 A0A0L7LVA7 A0A212F6P2 Q17F49 A0A1S4F521 B4NG99 A0A1I8Q2P6 A0A1I8N5F7 B4K7H0 A0A3B0JNY9 A0A182JJV1 B4GLN0 Q293T7 B4JI56 W5JR45 B3MTR5 B4M0W8 B4IB88 A0A182H7T3 Q9VEB4 A0A1W4U8U7 B4PL83 A0A1B0CTU3 A0A0L0BX74 A0A1L8D7Q0 B3NZ95 A0A182U5G8 A0A2C9H8J7 A0A182HZA8 Q7QAR2 A0A182MU92 A0A182KXI1 A0A182QNZ0 A0A182RZG6 A0A1B6CTF5 A0A182PS21 W8BXM5 A0A0A1WN33 A0A182VJJ5 A0A1A9UCL3 A0A1B0A628 A0A1A9X8P0 A0A1B0GEC2 A0A336MF22 A0A0K8VK14 A0A1B0BFD1 A0A182K3M2 A0A182Y7D8 A0A182VQ52 A0A1J1HUD3 A0A034WT76 A0A1Q3F2E9 A0A2J7Q1Z9 A0A2J7Q1Z1 B0XD92 A0A067R7T8 A0A2J7Q1Y6 Q17F48 A0A1S4F508 A0A1A9X216 A0A182NLH0 J9JXS7 A0A2S2QG36 A0A226E2A6 A0A1J1IF61 B4QUI0 A0A182FT12 A0A1D2N4P7 A0A293LR41 D6WKL7 A0A0A9WVV1 A0A0C9RB16 A0A1B6KIH5 A0A131YIM6 A0A224Z1C6 L7LUP4 T1HPB6 A0A026VU53 A0A131XAK6 A0A3L8E355
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion
Nucleus
Cytoplasm
Nucleus
Cytoplasm
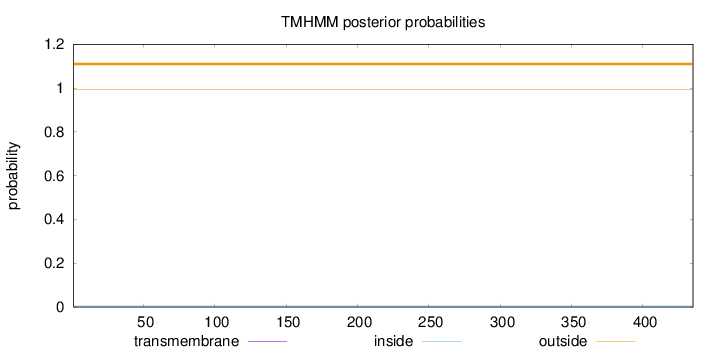
Length:
435
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00117
Exp number, first 60 AAs:
0.00095
Total prob of N-in:
0.00509
outside
1 - 435
Population Genetic Test Statistics
Pi
289.200755
Theta
192.84176
Tajima's D
1.56759
CLR
0.006405
CSRT
0.799310034498275
Interpretation
Uncertain
