Pre Gene Modal
BGIBMGA007804
Annotation
matrix_metalloproteinase_1_isoform_1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 0.962
Sequence
CDS
ATGTATTTAGCACAGTACGGTTATTTAAGTCCATCAGTAAGGAATCCTTCGAGTGGACACATTATGGATGAGAGCTCGTGGAGGAGAGCCATTGCCGAATTTCAGAGCTTCGCTGGTCTTAATGCTACGGGTGAACTGGATGACCAAACGAACGAAATGATGTCATTGCCAAGGTGTGGAGTTAGAGATAAAGTAGGCTTCGGAGAGAGTCGTGCCAAAAGATACGCATTGCAAGGGTCAAGATGGCGTGTCAAAAATCTGACGTACAAGATATCCAAGTACCCATCAAGACTGAATCGCGCCGAAGTCGATGCTGAGCTTGCCAAAGCCTTCTCCGTCTGGTCTGACTACACGGATCTTACTTTCACACAAAAGAGATCCGGTCAAGTCCACATAGAAATTAGGTTTGAGAAAGGCGAGCACGGTGACGGAGATCCTTTTGATGGTCCAGGAGGCACCCTCGCCCACGCTTATTTCCCGGTGTATGGAGGTGATGCCCATTTTGATGACGCTGAGATGTGGTCCATCAATTCCCGTAGAGGAACTAATCTTTTCCAGGTAGCAGCGCATGAGTTTGGTCACTCGCTGGGTCTCTCACACAGCGATGTAAGATCCGCATTGATGGCGCCGTTTTATCGTGGTTACGATCCTGCTTTCCAACTCGATCAGGACGACGTCCAAGGAATTCAGTCGCTTTATGGCCACAAGACCCAGACAGACATTGGTGGCGGTGGCGGCGGACTGATACCTTCCGTTCCTCGAGCGACCACGCAGCAGCCGTCAGCTGAGGATCCGGCTCTCTGCGCTGATCCTCGTATTGATACTATCTTTAATTCTGCCGACGGTTCGACTTTTGTATTCAAAGGCGATCATTACTGGAGGCTAACTGAAGATGGCGTAGCGGCTGGGTACCCGCGTCTTATTTCCCGAGCCTGGCCTGGTCTACCCGGAAACATTGATGCAGCATTTACCTACAAGAACGGCAAGACTTACTTCTTTAAGGGCTCGAAGTACTGGAGGTACAATGGGCAGAAGATGGACGGGGATTACCCCAAGGATATTAGTGAAGGCTTCACCGGTATCCCAGATAACCTAGATGCGGCTTTGGTTTGGTCCGGCAATGGCAAGATCTATTTCTACAAGGGCTCGAAATTCTGGCGATTTGACCCTGCACAACGGCCTCCCGTGAAAGCTACCTACCCGAAGCCTTTGTCTAACTGGGATGGAATACCTGATAACATCGACGCCGCCTTGCAGTACACTAACGGATACACCTACTTCTTCAAGGGGGGATCATACTGGAGATTCAACGACAGATTGTTCAGCGTGGACACAGACAACCCTCAGTTCCCGCGTTCAACCGCGTTCTGGTGGTTGGGCTGCAGCAGCGCGCCGCGCGGCACCGTCGGAGGTAACGCTCGACTATCAGACGACACTGTTCCTGCCGATGACGACGTCGGGGACATTACCTTCGACGCAGGTGTAAAATCATCCGCTCCCAGATCATTCTTCTGGTTCAGGAAATAG
Protein
MYLAQYGYLSPSVRNPSSGHIMDESSWRRAIAEFQSFAGLNATGELDDQTNEMMSLPRCGVRDKVGFGESRAKRYALQGSRWRVKNLTYKISKYPSRLNRAEVDAELAKAFSVWSDYTDLTFTQKRSGQVHIEIRFEKGEHGDGDPFDGPGGTLAHAYFPVYGGDAHFDDAEMWSINSRRGTNLFQVAAHEFGHSLGLSHSDVRSALMAPFYRGYDPAFQLDQDDVQGIQSLYGHKTQTDIGGGGGGLIPSVPRATTQQPSAEDPALCADPRIDTIFNSADGSTFVFKGDHYWRLTEDGVAAGYPRLISRAWPGLPGNIDAAFTYKNGKTYFFKGSKYWRYNGQKMDGDYPKDISEGFTGIPDNLDAALVWSGNGKIYFYKGSKFWRFDPAQRPPVKATYPKPLSNWDGIPDNIDAALQYTNGYTYFFKGGSYWRFNDRLFSVDTDNPQFPRSTAFWWLGCSSAPRGTVGGNARLSDDTVPADDDVGDITFDAGVKSSAPRSFFWFRK
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M10A family.
Uniprot
B2CI48
B2CI49
H9JE57
A0A2A4JFP2
G5CZ60
A0A2A4JF64
+ More
A0A2A4JEL5 A0A2A4JFL6 A0A2A4JE27 A0A437B5H5 A0A194R6V0 A0A212F4L3 A0A194PJU6 A0A1Q3FNC3 A0A1Q3FN05 A0A1Q3FN16 A0A1Q3FMU4 A0A1Q3FMS3 A0A1Q3FMY6 A0A1Q3FMU9 V5G6L1 A0A1Q3FMY0 A0A182QTZ7 A0A1Q3G4U6 A0A084WKS0 A0A1Q3G4T7 B0WWE1 A0A182N9E0 A0A182J688 A0A182K6X7 W8BIG6 A0A2C9H6V7 Q7QIJ4 A0A182G3J8 A0A2A3E310 A0A1I8MIR7 A0A182WWF5 A0A1I8MIR4 A0A182L3R1 A0A182TX16 A0A182I6N2 A0A182UZZ9 A0A1I8MIR8 A0A088AVW9 A0A310SIS7 A0A336MHW6 A0A1I8Q3R4 A0A0A1WMZ5 A0A034VPK7 A0A1I8MIR3 A0A336MF57 A0A034VPL4 A0A0K8UUG2 A0A0K8U7W3 T1PNC9 T1PDB0 Q179D2 A0A1I8MIR0 A0A0K8VXI9 A0A1B0CHM3 A0A034VL60 A0A1L8DPA9 A0A1L8DPG8 A0A0L0C2N4 A0A182FEG6 A0A1L8DPM9 A0A2M4CJD0 A0A182PIL9 A0A2M4CJ78 A0A2M3Z3D4 A0A182Y805 A0A2M4BJR9 A0A067XK10 A0A0L7QT13 A0A2A3E4T6 A0A0C9QPB4 A0A0C9QD22 A0A0C9REM2 D6WKX8 A0A1B0GCE8 A0A0R3NPE8 A0A0Q9XIN3 J9JXB6 A0A0R3NPP6 B4KTE1 A0A182MK74 A0A3B0JQP8 B4GGB5 A0A0J9UAM5 Q28XL6 A0A0Q9XEC7 A0A2H8TKH3 A0A0R3NVP4 A0A0B4JDA7 A0A0Q9XJV2 A0A1A9VT74 A0A0Q9X765 A0A3B0J335 A0A1B0BR59 A0A0Q9WE03
A0A2A4JEL5 A0A2A4JFL6 A0A2A4JE27 A0A437B5H5 A0A194R6V0 A0A212F4L3 A0A194PJU6 A0A1Q3FNC3 A0A1Q3FN05 A0A1Q3FN16 A0A1Q3FMU4 A0A1Q3FMS3 A0A1Q3FMY6 A0A1Q3FMU9 V5G6L1 A0A1Q3FMY0 A0A182QTZ7 A0A1Q3G4U6 A0A084WKS0 A0A1Q3G4T7 B0WWE1 A0A182N9E0 A0A182J688 A0A182K6X7 W8BIG6 A0A2C9H6V7 Q7QIJ4 A0A182G3J8 A0A2A3E310 A0A1I8MIR7 A0A182WWF5 A0A1I8MIR4 A0A182L3R1 A0A182TX16 A0A182I6N2 A0A182UZZ9 A0A1I8MIR8 A0A088AVW9 A0A310SIS7 A0A336MHW6 A0A1I8Q3R4 A0A0A1WMZ5 A0A034VPK7 A0A1I8MIR3 A0A336MF57 A0A034VPL4 A0A0K8UUG2 A0A0K8U7W3 T1PNC9 T1PDB0 Q179D2 A0A1I8MIR0 A0A0K8VXI9 A0A1B0CHM3 A0A034VL60 A0A1L8DPA9 A0A1L8DPG8 A0A0L0C2N4 A0A182FEG6 A0A1L8DPM9 A0A2M4CJD0 A0A182PIL9 A0A2M4CJ78 A0A2M3Z3D4 A0A182Y805 A0A2M4BJR9 A0A067XK10 A0A0L7QT13 A0A2A3E4T6 A0A0C9QPB4 A0A0C9QD22 A0A0C9REM2 D6WKX8 A0A1B0GCE8 A0A0R3NPE8 A0A0Q9XIN3 J9JXB6 A0A0R3NPP6 B4KTE1 A0A182MK74 A0A3B0JQP8 B4GGB5 A0A0J9UAM5 Q28XL6 A0A0Q9XEC7 A0A2H8TKH3 A0A0R3NVP4 A0A0B4JDA7 A0A0Q9XJV2 A0A1A9VT74 A0A0Q9X765 A0A3B0J335 A0A1B0BR59 A0A0Q9WE03
Pubmed
EMBL
EU496376
ACA64804.1
EU496377
ACA64805.1
BABH01002621
BABH01002622
+ More
NWSH01001761 PCG70222.1 JN415760 AEQ27775.1 PCG70214.1 PCG70216.1 PCG70220.1 PCG70221.1 RSAL01000152 RVE45722.1 KQ460615 KPJ13543.1 AGBW02010348 OWR48663.1 KQ459602 KPI93298.1 GFDL01006062 JAV28983.1 GFDL01006131 JAV28914.1 GFDL01006128 JAV28917.1 GFDL01006252 JAV28793.1 GFDL01006213 JAV28832.1 GFDL01006096 JAV28949.1 GFDL01006187 JAV28858.1 GALX01002746 JAB65720.1 GFDL01006157 JAV28888.1 AXCN02000560 GFDL01000220 JAV34825.1 ATLV01024134 KE525349 KFB50814.1 GFDL01000225 JAV34820.1 DS232144 EDS36033.1 GAMC01009754 GAMC01009753 GAMC01009752 JAB96803.1 AAAB01008807 EAA04040.3 EDO64756.1 EDO64757.1 JXUM01141621 KQ569279 KXJ68684.1 KZ288411 PBC26090.1 APCN01003684 KQ760792 OAD58997.1 UFQS01001123 UFQT01001123 SSX09120.1 SSX29031.1 GBXI01014070 JAD00222.1 GAKP01014900 GAKP01014891 JAC44052.1 SSX09121.1 SSX29032.1 GAKP01014895 JAC44057.1 GDHF01022022 GDHF01012453 GDHF01007959 JAI30292.1 JAI39861.1 JAI44355.1 GDHF01029631 GDHF01020114 GDHF01002913 JAI22683.1 JAI32200.1 JAI49401.1 KA649448 AFP64077.1 KA646105 AFP60734.1 CH477354 EAT42838.1 GDHF01008701 JAI43613.1 AJWK01012495 AJWK01012496 AJWK01012497 AJWK01012498 GAKP01014896 JAC44056.1 GFDF01005782 JAV08302.1 GFDF01005793 JAV08291.1 JRES01000975 KNC26613.1 GFDF01005794 JAV08290.1 GGFL01001235 MBW65413.1 GGFL01001234 MBW65412.1 GGFM01002276 MBW23027.1 GGFJ01004060 MBW53201.1 JX399873 AGI44421.1 KQ414755 KOC61694.1 PBC26091.1 GBYB01005434 JAG75201.1 GBYB01001214 GBYB01005435 JAG70981.1 JAG75202.1 GBYB01005436 JAG75203.1 KQ971343 EFA04667.1 CCAG010007850 CCAG010007851 CM000071 KRT02977.1 CH933808 KRG04151.1 ABLF02036098 KRT02978.1 EDW08502.1 KRG04153.1 AXCM01006016 OUUW01000001 SPP75686.1 CH479183 EDW35535.1 CM002911 KMY96410.1 EAL26300.3 KRT02980.1 KRG04154.1 GFXV01002839 MBW14644.1 KRT02976.1 AE013599 ADV37248.1 KRG04152.1 KRG04150.1 SPP75685.1 JXJN01018910 CH940648 KRF79195.1
NWSH01001761 PCG70222.1 JN415760 AEQ27775.1 PCG70214.1 PCG70216.1 PCG70220.1 PCG70221.1 RSAL01000152 RVE45722.1 KQ460615 KPJ13543.1 AGBW02010348 OWR48663.1 KQ459602 KPI93298.1 GFDL01006062 JAV28983.1 GFDL01006131 JAV28914.1 GFDL01006128 JAV28917.1 GFDL01006252 JAV28793.1 GFDL01006213 JAV28832.1 GFDL01006096 JAV28949.1 GFDL01006187 JAV28858.1 GALX01002746 JAB65720.1 GFDL01006157 JAV28888.1 AXCN02000560 GFDL01000220 JAV34825.1 ATLV01024134 KE525349 KFB50814.1 GFDL01000225 JAV34820.1 DS232144 EDS36033.1 GAMC01009754 GAMC01009753 GAMC01009752 JAB96803.1 AAAB01008807 EAA04040.3 EDO64756.1 EDO64757.1 JXUM01141621 KQ569279 KXJ68684.1 KZ288411 PBC26090.1 APCN01003684 KQ760792 OAD58997.1 UFQS01001123 UFQT01001123 SSX09120.1 SSX29031.1 GBXI01014070 JAD00222.1 GAKP01014900 GAKP01014891 JAC44052.1 SSX09121.1 SSX29032.1 GAKP01014895 JAC44057.1 GDHF01022022 GDHF01012453 GDHF01007959 JAI30292.1 JAI39861.1 JAI44355.1 GDHF01029631 GDHF01020114 GDHF01002913 JAI22683.1 JAI32200.1 JAI49401.1 KA649448 AFP64077.1 KA646105 AFP60734.1 CH477354 EAT42838.1 GDHF01008701 JAI43613.1 AJWK01012495 AJWK01012496 AJWK01012497 AJWK01012498 GAKP01014896 JAC44056.1 GFDF01005782 JAV08302.1 GFDF01005793 JAV08291.1 JRES01000975 KNC26613.1 GFDF01005794 JAV08290.1 GGFL01001235 MBW65413.1 GGFL01001234 MBW65412.1 GGFM01002276 MBW23027.1 GGFJ01004060 MBW53201.1 JX399873 AGI44421.1 KQ414755 KOC61694.1 PBC26091.1 GBYB01005434 JAG75201.1 GBYB01001214 GBYB01005435 JAG70981.1 JAG75202.1 GBYB01005436 JAG75203.1 KQ971343 EFA04667.1 CCAG010007850 CCAG010007851 CM000071 KRT02977.1 CH933808 KRG04151.1 ABLF02036098 KRT02978.1 EDW08502.1 KRG04153.1 AXCM01006016 OUUW01000001 SPP75686.1 CH479183 EDW35535.1 CM002911 KMY96410.1 EAL26300.3 KRT02980.1 KRG04154.1 GFXV01002839 MBW14644.1 KRT02976.1 AE013599 ADV37248.1 KRG04152.1 KRG04150.1 SPP75685.1 JXJN01018910 CH940648 KRF79195.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
+ More
UP000075886 UP000030765 UP000002320 UP000075884 UP000075880 UP000075881 UP000076407 UP000007062 UP000069940 UP000249989 UP000242457 UP000095301 UP000075882 UP000075902 UP000075840 UP000075903 UP000005203 UP000095300 UP000008820 UP000092461 UP000037069 UP000069272 UP000075885 UP000076408 UP000053825 UP000007266 UP000092444 UP000001819 UP000009192 UP000007819 UP000075883 UP000268350 UP000008744 UP000000803 UP000078200 UP000092460 UP000008792
UP000075886 UP000030765 UP000002320 UP000075884 UP000075880 UP000075881 UP000076407 UP000007062 UP000069940 UP000249989 UP000242457 UP000095301 UP000075882 UP000075902 UP000075840 UP000075903 UP000005203 UP000095300 UP000008820 UP000092461 UP000037069 UP000069272 UP000075885 UP000076408 UP000053825 UP000007266 UP000092444 UP000001819 UP000009192 UP000007819 UP000075883 UP000268350 UP000008744 UP000000803 UP000078200 UP000092460 UP000008792
Interpro
IPR006026
Peptidase_Metallo
+ More
IPR036365 PGBD-like_sf
IPR000585 Hemopexin-like_dom
IPR018487 Hemopexin-like_repeat
IPR036375 Hemopexin-like_dom_sf
IPR033739 M10A_MMP
IPR021190 Pept_M10A
IPR018486 Hemopexin_CS
IPR024079 MetalloPept_cat_dom_sf
IPR002477 Peptidoglycan-bd-like
IPR001818 Pept_M10_metallopeptidase
IPR036365 PGBD-like_sf
IPR000585 Hemopexin-like_dom
IPR018487 Hemopexin-like_repeat
IPR036375 Hemopexin-like_dom_sf
IPR033739 M10A_MMP
IPR021190 Pept_M10A
IPR018486 Hemopexin_CS
IPR024079 MetalloPept_cat_dom_sf
IPR002477 Peptidoglycan-bd-like
IPR001818 Pept_M10_metallopeptidase
Gene 3D
ProteinModelPortal
B2CI48
B2CI49
H9JE57
A0A2A4JFP2
G5CZ60
A0A2A4JF64
+ More
A0A2A4JEL5 A0A2A4JFL6 A0A2A4JE27 A0A437B5H5 A0A194R6V0 A0A212F4L3 A0A194PJU6 A0A1Q3FNC3 A0A1Q3FN05 A0A1Q3FN16 A0A1Q3FMU4 A0A1Q3FMS3 A0A1Q3FMY6 A0A1Q3FMU9 V5G6L1 A0A1Q3FMY0 A0A182QTZ7 A0A1Q3G4U6 A0A084WKS0 A0A1Q3G4T7 B0WWE1 A0A182N9E0 A0A182J688 A0A182K6X7 W8BIG6 A0A2C9H6V7 Q7QIJ4 A0A182G3J8 A0A2A3E310 A0A1I8MIR7 A0A182WWF5 A0A1I8MIR4 A0A182L3R1 A0A182TX16 A0A182I6N2 A0A182UZZ9 A0A1I8MIR8 A0A088AVW9 A0A310SIS7 A0A336MHW6 A0A1I8Q3R4 A0A0A1WMZ5 A0A034VPK7 A0A1I8MIR3 A0A336MF57 A0A034VPL4 A0A0K8UUG2 A0A0K8U7W3 T1PNC9 T1PDB0 Q179D2 A0A1I8MIR0 A0A0K8VXI9 A0A1B0CHM3 A0A034VL60 A0A1L8DPA9 A0A1L8DPG8 A0A0L0C2N4 A0A182FEG6 A0A1L8DPM9 A0A2M4CJD0 A0A182PIL9 A0A2M4CJ78 A0A2M3Z3D4 A0A182Y805 A0A2M4BJR9 A0A067XK10 A0A0L7QT13 A0A2A3E4T6 A0A0C9QPB4 A0A0C9QD22 A0A0C9REM2 D6WKX8 A0A1B0GCE8 A0A0R3NPE8 A0A0Q9XIN3 J9JXB6 A0A0R3NPP6 B4KTE1 A0A182MK74 A0A3B0JQP8 B4GGB5 A0A0J9UAM5 Q28XL6 A0A0Q9XEC7 A0A2H8TKH3 A0A0R3NVP4 A0A0B4JDA7 A0A0Q9XJV2 A0A1A9VT74 A0A0Q9X765 A0A3B0J335 A0A1B0BR59 A0A0Q9WE03
A0A2A4JEL5 A0A2A4JFL6 A0A2A4JE27 A0A437B5H5 A0A194R6V0 A0A212F4L3 A0A194PJU6 A0A1Q3FNC3 A0A1Q3FN05 A0A1Q3FN16 A0A1Q3FMU4 A0A1Q3FMS3 A0A1Q3FMY6 A0A1Q3FMU9 V5G6L1 A0A1Q3FMY0 A0A182QTZ7 A0A1Q3G4U6 A0A084WKS0 A0A1Q3G4T7 B0WWE1 A0A182N9E0 A0A182J688 A0A182K6X7 W8BIG6 A0A2C9H6V7 Q7QIJ4 A0A182G3J8 A0A2A3E310 A0A1I8MIR7 A0A182WWF5 A0A1I8MIR4 A0A182L3R1 A0A182TX16 A0A182I6N2 A0A182UZZ9 A0A1I8MIR8 A0A088AVW9 A0A310SIS7 A0A336MHW6 A0A1I8Q3R4 A0A0A1WMZ5 A0A034VPK7 A0A1I8MIR3 A0A336MF57 A0A034VPL4 A0A0K8UUG2 A0A0K8U7W3 T1PNC9 T1PDB0 Q179D2 A0A1I8MIR0 A0A0K8VXI9 A0A1B0CHM3 A0A034VL60 A0A1L8DPA9 A0A1L8DPG8 A0A0L0C2N4 A0A182FEG6 A0A1L8DPM9 A0A2M4CJD0 A0A182PIL9 A0A2M4CJ78 A0A2M3Z3D4 A0A182Y805 A0A2M4BJR9 A0A067XK10 A0A0L7QT13 A0A2A3E4T6 A0A0C9QPB4 A0A0C9QD22 A0A0C9REM2 D6WKX8 A0A1B0GCE8 A0A0R3NPE8 A0A0Q9XIN3 J9JXB6 A0A0R3NPP6 B4KTE1 A0A182MK74 A0A3B0JQP8 B4GGB5 A0A0J9UAM5 Q28XL6 A0A0Q9XEC7 A0A2H8TKH3 A0A0R3NVP4 A0A0B4JDA7 A0A0Q9XJV2 A0A1A9VT74 A0A0Q9X765 A0A3B0J335 A0A1B0BR59 A0A0Q9WE03
PDB
1SU3
E-value=5.85001e-59,
Score=578
Ontologies
GO
GO:0031012
GO:0004222
GO:0008270
GO:0016021
GO:0005615
GO:0030198
GO:0030574
GO:0007591
GO:0007419
GO:0003144
GO:0035001
GO:0035159
GO:0042246
GO:0002168
GO:0007505
GO:0030425
GO:0034769
GO:0097156
GO:0007561
GO:0045216
GO:0007155
GO:0007424
GO:0042060
GO:0071711
GO:0006508
GO:0005509
GO:0008237
GO:0043565
GO:0016876
GO:0043039
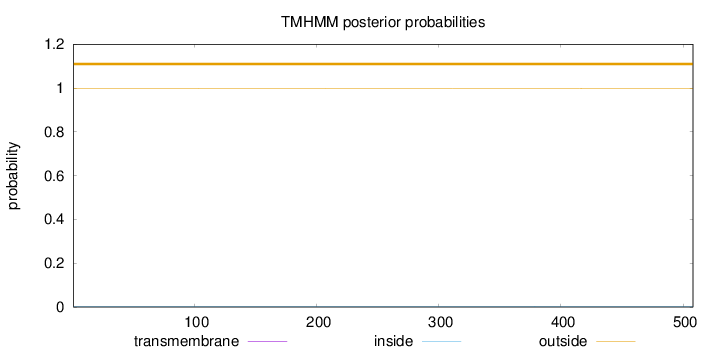
Topology
Length:
508
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00171
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00171
outside
1 - 508
Population Genetic Test Statistics
Pi
224.882217
Theta
184.637302
Tajima's D
0.645259
CLR
0.177513
CSRT
0.555722213889306
Interpretation
Uncertain
