Gene
KWMTBOMO08935
Annotation
PREDICTED:_lipopolysaccharide-induced_tumor_necrosis_factor-alpha_factor_homolog_[Bombyx_mori]
Transcription factor
Location in the cell
Extracellular Reliability : 1.523 PlasmaMembrane Reliability : 1.529
Sequence
CDS
ATGGATCTTCTACAAGTAGGCTCGGAGCCTGTAGGCATGAGGTGCCCTTATTGCCAAGAAGACATAATGACGAGGGCTATTTACAAAAATTCGACCATCACTCATATCGTTGCTGCAGTGCTGGGTGTATTGTTCTGGTGGCTGTGTTGCTGTATAATTCCGTATACAACGAAACGTTGGAAAAACGTGGAGCATAATTGTCCCAATTGTCATAAATTCCTGGGTGTCTATGAAAGAACGAGAGTGATTTAA
Protein
MDLLQVGSEPVGMRCPYCQEDIMTRAIYKNSTITHIVAAVLGVLFWWLCCCIIPYTTKRWKNVEHNCPNCHKFLGVYERTRVI
Summary
Uniprot
A0A1E1WBQ9
A0A194R6U3
A0A2H1W2L1
A0A2A4JR63
A0A437ATJ0
A0A2H1WAD5
+ More
A0A2W1BNA3 A0A2B4RVD9 A0A3M6UJR4 A0A1B0BU36 A0A0K8UTC2 A0A2A4JVU8 A0A437BIX7 A0A3M6TLJ8 A0A2B4SN90 A0A336MY04 A0A194ANP1 A0A336MTF6 A0A0C2GNG4 S4P885 A0A2J7RKB4 A0A2I4CT57 A0A1S4FG64 Q172M2 A0A182G7S7 A0A2A4JW93 B4KP83 A9XBI8 A0A0S1TPH1 A0A212FFW2 B4QHK9 K1R4U6 G9FXD7 A0A437BJ34 B4I887 A0A1S4FG68 K1QAE1 A0A1J1ILL1 B4P5J4 D2KK68 A8DYM6 A0A3Q3MAI5 A0A3Q1G347 W5XM22 A0A0J9RK55 A0A0B1TWW6 A0A0B1THI0 A0A3P7JR87 A0A194PJC2 B3FMU5 A0A3B4AL09 A0A023EH32 A0A3Q3QD55 A0A182GCI1 Q172M1 A0A2W1BQA8 F1BZ88 A0A194RSS6 G1FET2 A0A3Q3EZX7 A0A3R7QXD1 K1QR64 B3NLH2 A0A3Q2PHE7 A0A3Q0KGK6 A0A1W4VQJ0 A0A1L8EIY1 A0A194R8G0 G4VQL6 A0A2H1WJ10 A0A1S3IHN4 A8CA04 K1RPA3 A0A182JDM5 A0A2W1BN81 A0A194PQ83 B0X8G2 W5XM11 A0A3Q2D6L8 A0A1S3IIP9 W2TR24 I4DPT9 A0A067RLV9 A0A182JK51 A0A1S3II01 A0A084VYA4 A0A1B0GQM0 A0A0M4EEI3 A0A182PVK6 A0A210QGT7 A0A1Q3FKN6 A0A182VL92 T1DFW1 A0A034WCJ7 A0A0C9QIY8 A0A182TGD0 A0A182KYN3 A7UV77 A0A182HZ06
A0A2W1BNA3 A0A2B4RVD9 A0A3M6UJR4 A0A1B0BU36 A0A0K8UTC2 A0A2A4JVU8 A0A437BIX7 A0A3M6TLJ8 A0A2B4SN90 A0A336MY04 A0A194ANP1 A0A336MTF6 A0A0C2GNG4 S4P885 A0A2J7RKB4 A0A2I4CT57 A0A1S4FG64 Q172M2 A0A182G7S7 A0A2A4JW93 B4KP83 A9XBI8 A0A0S1TPH1 A0A212FFW2 B4QHK9 K1R4U6 G9FXD7 A0A437BJ34 B4I887 A0A1S4FG68 K1QAE1 A0A1J1ILL1 B4P5J4 D2KK68 A8DYM6 A0A3Q3MAI5 A0A3Q1G347 W5XM22 A0A0J9RK55 A0A0B1TWW6 A0A0B1THI0 A0A3P7JR87 A0A194PJC2 B3FMU5 A0A3B4AL09 A0A023EH32 A0A3Q3QD55 A0A182GCI1 Q172M1 A0A2W1BQA8 F1BZ88 A0A194RSS6 G1FET2 A0A3Q3EZX7 A0A3R7QXD1 K1QR64 B3NLH2 A0A3Q2PHE7 A0A3Q0KGK6 A0A1W4VQJ0 A0A1L8EIY1 A0A194R8G0 G4VQL6 A0A2H1WJ10 A0A1S3IHN4 A8CA04 K1RPA3 A0A182JDM5 A0A2W1BN81 A0A194PQ83 B0X8G2 W5XM11 A0A3Q2D6L8 A0A1S3IIP9 W2TR24 I4DPT9 A0A067RLV9 A0A182JK51 A0A1S3II01 A0A084VYA4 A0A1B0GQM0 A0A0M4EEI3 A0A182PVK6 A0A210QGT7 A0A1Q3FKN6 A0A182VL92 T1DFW1 A0A034WCJ7 A0A0C9QIY8 A0A182TGD0 A0A182KYN3 A7UV77 A0A182HZ06
Pubmed
26354079
28756777
30382153
23622113
17510324
26483478
+ More
17994087 17980621 22118469 22936249 22992520 22138218 17550304 21567197 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18395262 25463417 24945155 21736897 22253936 17760985 22651552 24845553 24438588 28812685 24330624 25348373 20966253 12364791 14747013 17210077
17994087 17980621 22118469 22936249 22992520 22138218 17550304 21567197 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18395262 25463417 24945155 21736897 22253936 17760985 22651552 24845553 24438588 28812685 24330624 25348373 20966253 12364791 14747013 17210077
EMBL
GDQN01006626
GDQN01002211
GDQN01001780
JAT84428.1
JAT88843.1
JAT89274.1
+ More
KQ460615 KPJ13538.1 ODYU01005917 SOQ47311.1 NWSH01000840 PCG73912.1 RSAL01000488 RVE41554.1 ODYU01007335 SOQ50013.1 KZ149983 PZC75751.1 LSMT01000314 PFX20520.1 RCHS01001413 RMX53618.1 JXJN01020464 JXJN01020465 GDHF01022493 JAI29821.1 NWSH01000518 PCG75906.1 RSAL01000049 RVE50382.1 RCHS01003379 RMX42283.1 LSMT01000049 PFX30533.1 UFQT01003106 SSX34585.1 GELH01000160 GELH01000159 JAS04112.1 UFQT01002583 SSX33712.1 KN729055 KIH62825.1 GAIX01006056 JAA86504.1 NEVH01002981 PNF41277.1 CH477434 EAT40986.1 JXUM01046825 JXUM01054329 KQ561816 KQ561492 KXJ77441.1 KXJ78397.1 PCG75904.1 CH933808 EDW09059.1 EF157939 EF157940 ABO70331.1 KT023069 ALM25911.1 AGBW02008757 OWR52625.1 CM000362 CM002911 EDX08238.1 KMY95840.1 JH817566 EKC36235.1 JN216844 AEX08893.1 RVE50383.1 CH480824 EDW56812.1 JH816790 EKC33607.1 CVRI01000055 CRL01127.1 CM000158 EDW91825.1 KRK00128.1 GU217721 ADA77533.1 AE013599 ABV53888.2 AHN56535.1 KF051277 AHI17306.1 KMY95839.1 KN549200 KHJ99985.1 KN550019 KHJ95272.1 UYYB01146756 VDM85886.1 KQ459606 KPI91185.1 EU249541 ACA96822.1 GAPW01005437 JAC08161.1 JXUM01054326 JXUM01054327 JXUM01054328 KXJ77440.1 EAT40987.1 PZC75755.1 HQ174259 ADX31291.1 KQ459700 KPJ20400.1 JN180640 AEK86526.1 QCYY01000928 ROT81727.1 EKC36233.1 CH954179 EDV54888.1 GFDG01000195 JAV18604.1 KPJ13540.1 HE601631 CCD82576.1 ODYU01008993 SOQ53063.1 EU035828 ABV44730.1 EKC36231.1 KZ150055 PZC74326.1 KQ459602 KPI93295.1 DS232488 EDS42498.1 KF110681 AHI17296.1 KI658196 ETN83482.1 AK403777 BAM19929.1 KK852598 KDR20553.1 ATLV01018349 KE525231 KFB42948.1 AJVK01017302 CP012524 ALC42502.1 NEDP02003747 OWF47962.1 GFDL01006896 JAV28149.1 GALA01000412 JAA94440.1 GAKP01006543 JAC52409.1 GBYB01003504 JAG73271.1 AAAB01008984 EDO63389.1 APCN01005837
KQ460615 KPJ13538.1 ODYU01005917 SOQ47311.1 NWSH01000840 PCG73912.1 RSAL01000488 RVE41554.1 ODYU01007335 SOQ50013.1 KZ149983 PZC75751.1 LSMT01000314 PFX20520.1 RCHS01001413 RMX53618.1 JXJN01020464 JXJN01020465 GDHF01022493 JAI29821.1 NWSH01000518 PCG75906.1 RSAL01000049 RVE50382.1 RCHS01003379 RMX42283.1 LSMT01000049 PFX30533.1 UFQT01003106 SSX34585.1 GELH01000160 GELH01000159 JAS04112.1 UFQT01002583 SSX33712.1 KN729055 KIH62825.1 GAIX01006056 JAA86504.1 NEVH01002981 PNF41277.1 CH477434 EAT40986.1 JXUM01046825 JXUM01054329 KQ561816 KQ561492 KXJ77441.1 KXJ78397.1 PCG75904.1 CH933808 EDW09059.1 EF157939 EF157940 ABO70331.1 KT023069 ALM25911.1 AGBW02008757 OWR52625.1 CM000362 CM002911 EDX08238.1 KMY95840.1 JH817566 EKC36235.1 JN216844 AEX08893.1 RVE50383.1 CH480824 EDW56812.1 JH816790 EKC33607.1 CVRI01000055 CRL01127.1 CM000158 EDW91825.1 KRK00128.1 GU217721 ADA77533.1 AE013599 ABV53888.2 AHN56535.1 KF051277 AHI17306.1 KMY95839.1 KN549200 KHJ99985.1 KN550019 KHJ95272.1 UYYB01146756 VDM85886.1 KQ459606 KPI91185.1 EU249541 ACA96822.1 GAPW01005437 JAC08161.1 JXUM01054326 JXUM01054327 JXUM01054328 KXJ77440.1 EAT40987.1 PZC75755.1 HQ174259 ADX31291.1 KQ459700 KPJ20400.1 JN180640 AEK86526.1 QCYY01000928 ROT81727.1 EKC36233.1 CH954179 EDV54888.1 GFDG01000195 JAV18604.1 KPJ13540.1 HE601631 CCD82576.1 ODYU01008993 SOQ53063.1 EU035828 ABV44730.1 EKC36231.1 KZ150055 PZC74326.1 KQ459602 KPI93295.1 DS232488 EDS42498.1 KF110681 AHI17296.1 KI658196 ETN83482.1 AK403777 BAM19929.1 KK852598 KDR20553.1 ATLV01018349 KE525231 KFB42948.1 AJVK01017302 CP012524 ALC42502.1 NEDP02003747 OWF47962.1 GFDL01006896 JAV28149.1 GALA01000412 JAA94440.1 GAKP01006543 JAC52409.1 GBYB01003504 JAG73271.1 AAAB01008984 EDO63389.1 APCN01005837
Proteomes
UP000053240
UP000218220
UP000283053
UP000225706
UP000275408
UP000092460
+ More
UP000235965 UP000192220 UP000008820 UP000069940 UP000249989 UP000009192 UP000007151 UP000000304 UP000005408 UP000001292 UP000183832 UP000002282 UP000000803 UP000261640 UP000257200 UP000053268 UP000261520 UP000261600 UP000261660 UP000283509 UP000008711 UP000265000 UP000008854 UP000192221 UP000085678 UP000075880 UP000002320 UP000265020 UP000027135 UP000030765 UP000092462 UP000092553 UP000075885 UP000242188 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840
UP000235965 UP000192220 UP000008820 UP000069940 UP000249989 UP000009192 UP000007151 UP000000304 UP000005408 UP000001292 UP000183832 UP000002282 UP000000803 UP000261640 UP000257200 UP000053268 UP000261520 UP000261600 UP000261660 UP000283509 UP000008711 UP000265000 UP000008854 UP000192221 UP000085678 UP000075880 UP000002320 UP000265020 UP000027135 UP000030765 UP000092462 UP000092553 UP000075885 UP000242188 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840
Pfam
PF10601 zf-LITAF-like
ProteinModelPortal
A0A1E1WBQ9
A0A194R6U3
A0A2H1W2L1
A0A2A4JR63
A0A437ATJ0
A0A2H1WAD5
+ More
A0A2W1BNA3 A0A2B4RVD9 A0A3M6UJR4 A0A1B0BU36 A0A0K8UTC2 A0A2A4JVU8 A0A437BIX7 A0A3M6TLJ8 A0A2B4SN90 A0A336MY04 A0A194ANP1 A0A336MTF6 A0A0C2GNG4 S4P885 A0A2J7RKB4 A0A2I4CT57 A0A1S4FG64 Q172M2 A0A182G7S7 A0A2A4JW93 B4KP83 A9XBI8 A0A0S1TPH1 A0A212FFW2 B4QHK9 K1R4U6 G9FXD7 A0A437BJ34 B4I887 A0A1S4FG68 K1QAE1 A0A1J1ILL1 B4P5J4 D2KK68 A8DYM6 A0A3Q3MAI5 A0A3Q1G347 W5XM22 A0A0J9RK55 A0A0B1TWW6 A0A0B1THI0 A0A3P7JR87 A0A194PJC2 B3FMU5 A0A3B4AL09 A0A023EH32 A0A3Q3QD55 A0A182GCI1 Q172M1 A0A2W1BQA8 F1BZ88 A0A194RSS6 G1FET2 A0A3Q3EZX7 A0A3R7QXD1 K1QR64 B3NLH2 A0A3Q2PHE7 A0A3Q0KGK6 A0A1W4VQJ0 A0A1L8EIY1 A0A194R8G0 G4VQL6 A0A2H1WJ10 A0A1S3IHN4 A8CA04 K1RPA3 A0A182JDM5 A0A2W1BN81 A0A194PQ83 B0X8G2 W5XM11 A0A3Q2D6L8 A0A1S3IIP9 W2TR24 I4DPT9 A0A067RLV9 A0A182JK51 A0A1S3II01 A0A084VYA4 A0A1B0GQM0 A0A0M4EEI3 A0A182PVK6 A0A210QGT7 A0A1Q3FKN6 A0A182VL92 T1DFW1 A0A034WCJ7 A0A0C9QIY8 A0A182TGD0 A0A182KYN3 A7UV77 A0A182HZ06
A0A2W1BNA3 A0A2B4RVD9 A0A3M6UJR4 A0A1B0BU36 A0A0K8UTC2 A0A2A4JVU8 A0A437BIX7 A0A3M6TLJ8 A0A2B4SN90 A0A336MY04 A0A194ANP1 A0A336MTF6 A0A0C2GNG4 S4P885 A0A2J7RKB4 A0A2I4CT57 A0A1S4FG64 Q172M2 A0A182G7S7 A0A2A4JW93 B4KP83 A9XBI8 A0A0S1TPH1 A0A212FFW2 B4QHK9 K1R4U6 G9FXD7 A0A437BJ34 B4I887 A0A1S4FG68 K1QAE1 A0A1J1ILL1 B4P5J4 D2KK68 A8DYM6 A0A3Q3MAI5 A0A3Q1G347 W5XM22 A0A0J9RK55 A0A0B1TWW6 A0A0B1THI0 A0A3P7JR87 A0A194PJC2 B3FMU5 A0A3B4AL09 A0A023EH32 A0A3Q3QD55 A0A182GCI1 Q172M1 A0A2W1BQA8 F1BZ88 A0A194RSS6 G1FET2 A0A3Q3EZX7 A0A3R7QXD1 K1QR64 B3NLH2 A0A3Q2PHE7 A0A3Q0KGK6 A0A1W4VQJ0 A0A1L8EIY1 A0A194R8G0 G4VQL6 A0A2H1WJ10 A0A1S3IHN4 A8CA04 K1RPA3 A0A182JDM5 A0A2W1BN81 A0A194PQ83 B0X8G2 W5XM11 A0A3Q2D6L8 A0A1S3IIP9 W2TR24 I4DPT9 A0A067RLV9 A0A182JK51 A0A1S3II01 A0A084VYA4 A0A1B0GQM0 A0A0M4EEI3 A0A182PVK6 A0A210QGT7 A0A1Q3FKN6 A0A182VL92 T1DFW1 A0A034WCJ7 A0A0C9QIY8 A0A182TGD0 A0A182KYN3 A7UV77 A0A182HZ06
Ontologies
PANTHER
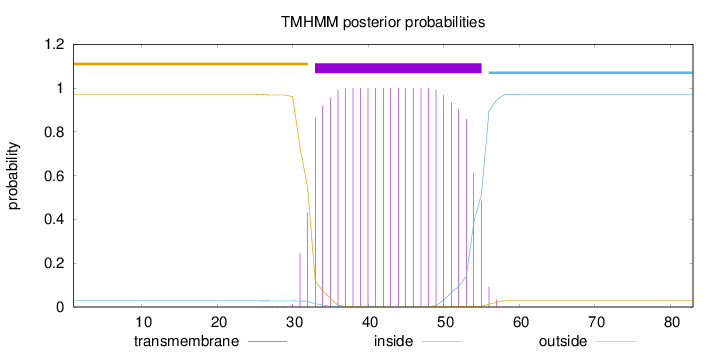
Topology
Length:
83
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.29921
Exp number, first 60 AAs:
22.2992
Total prob of N-in:
0.02845
POSSIBLE N-term signal
sequence
outside
1 - 32
TMhelix
33 - 55
inside
56 - 83
Population Genetic Test Statistics
Pi
210.417013
Theta
153.438365
Tajima's D
1.280749
CLR
0
CSRT
0.739763011849408
Interpretation
Uncertain
