Pre Gene Modal
BGIBMGA007806
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_integrator_complex_subunit_8_[Bombyx_mori]
Full name
Mothers against decapentaplegic homolog
+ More
Integrator complex subunit 8
Integrator complex subunit 8
Alternative Name
SMAD family member
Location in the cell
Cytoplasmic Reliability : 1.449
Sequence
CDS
ATGGATGTAGATCTGTTAAGACCCGGCACTGTGCCCATTTCACCTGACACTATATTATGGTTCGAGTTTCTCTTACATCCAGAACTGCTAAAGAATCATCTCAGCAAACCTAACCCAGAGCCATCAGCATGTGAACTTATAGAGGAATTTTTGTCAGTTGACTCCAAGGGTGCAAACTCGAAGCCTACTAGTGAACGAATACCAGATACTGATGCTCCACCCACCCCACCATCGGCGATTTCATCTCCCATACAGTTTACTCGAAAACAACTGGCTTTGAAAATCCTTGGACTGAAAGTGGCAGCATCATTACATTGGAACCTAGATACTTTGGAATTGAATCTAGCACCTCAAATACAGCACCAACTCATGCAAGACCTTGTTTACATGGCCACAGATGCAGGTTTTGCTGTACCGCCACAGGAAATACCAGCAGAGTTAATGCAAAAGCCACAAGTTATATTTGCATTAACTCTATACCACAGATGGACATTGAGATTCCCTGTGAAGGCGGCTTTGTATGCTAAATCATCAAAAATATACATGCACATGCCTGGAATGCAGACAGATGGTGGCTACCCTCCATTGAACCAAAATTTCGAAAAAATATTACGTATGTGTGAAGGAAATCGTACACATAGTATAAAATTTCTCGAAGAAGTTTTGTCCATGTACGAACTCACGTCAAATTCTAGCCGTAGAGAAACTAAAAAAATTAAAGTTCCGCCAATGGAGGCATTTATTCATCTCACTGAAGATTCTGATGAAGTAATACATAACTGGGATGCTGGAGACATTCTAATTACACAATTTGAATTAGCGATGCAAATACACTTTGATCTTTGTTACAATTATTTTTTCTATGGACAACATGATTTAGCAAAAAAACATATACTTGGATGCAGAGAAAATTCTAACCTTTTAGAAAGAGAAGTGTCTATCTATGGTTATGGTCCACACAAAGTTGTTCCTTGGGGTGAATTTTATTATGCTAGTATGACTAAAGATGATATTATTGGTTATATCAGAGCCCTCAACTTAGGTCAAGAAGTTTTAAATGAAGAAGCATCGTTATTGCAAAAACTTCAAGAATCTATAGCTAATCATTATACGGGCATCATTGCAGTTTTACAGGCTGATAATTTAGCTCGTGAAATTCCGATGATTCACAGACAAGTCGTCGAGTTGGATATACAAGGATCAGCGTCAAGTGGTGCATTCACAGTAGCCAGAGATCTCCTTAATCGGGTATCGGCTCTGAATGCAGTTAGATATGCTCTCGAAGGAGGCATACCCTCTACACATCCAGATTTCTTGAATAAATTCAAAACAGTTGGCATAAAGTTTTTCGATCTCCTCTTTTGGGCTCTTGGACCAGTATTACTATCGGATCTTTCTTTAGAAGACTGGCAACACTTACGAATGTTTTTCTTACATTTGATAACCTCTCAATGCAGACTTCCAATCGATAGAATAGATGAGTATATGAAAAAACATATTGGTGAAGCAGCAGGAAATATAAGAAAAAAACTGATTACCGATGAGCAACTTAGCATAATTCTGAACGATCCAATTAACAACGACGATGAAAACATGGAAATTCCTCGGGAACTACTCACCGACGACTGGGAAACGCCTGATTTTGATTTCAAAACTGTCCCTGAATTGGACATGGGCAGATTAAAGAAACGTCTCATCGAAGCTTCGACGGCTGATGACGTGCGCATGTGTCTAGTGAAACTAGCAATGATGTCTCCTTCGTTGCCTTTGTGGAAGTTAAGCCCGTCGTGGAAACCGGCAGGTGTATTGGGAAATGCTTTGATGTCGCTTAACAGAGGATTTTTGCAAGATTTTGGCTATGTAGTCTCGGGCGCAGCCCGTGCTCGTGCCGAATCGGGATCTGCACGAACTGCGTTGTCTCTCTTGTCGGTGCTTGAAGGAGAAGCTCGTAGTCAACTAGGCGGTGGCACTGATCCAGTGCTGTATCGACTGTGCCGACAGTTATCGTGGGAAGTACTGCTGCTGCAAGTCAACGTGATGCTCTCCGAGTGGCCTCACCATAGACTCAACCTTACTGCACTGGCAAATAAATGCAAAGCTTGCATTGCAGCGGCTAATTCTGGTGACGGTATCATACCACGACCACAGGTTTTAGAAGCGTGCTGGAGTTGTTTGGTTAATTCTTGCGAATGGGAAGGTCTGGGTGTCACCACAGGCCCTGGAGAATTAGCTTCCGCTCTTTGTGCGGCATGCTTTGAACTACAACGCGGTAAAGGCTCTAGAAAATTTCCAAGACCTCTATGGGACTACGCGTTATCGGTGTACAGTAACGGCCCAAATATTTCAATGAAACGTAGCGCGGGAGGGATGTCCCATGCGCGTGAAATGATGAACGCGGCGGCTGAGTCGCGCAACGCGTTCAACGGCTTCCTGGCAACCCTGCGCGAGCCACTCGCGATCAGCATCATGATGTCGCTCTTGGCTAGAATACACAACCTGCTGATCGACGACAACTCGCTCGAACTCAACACCGAATACACGAGTTTGTGGCCTTCTAGCGTTTCGAACATAAACAATTACAACATAAAATTCGTGTTAGAATCTCTAACCGATATGTTAGAAAGGAGTCTCAAATTATACCCCTACAATACTTCGTGGTGA
Protein
MDVDLLRPGTVPISPDTILWFEFLLHPELLKNHLSKPNPEPSACELIEEFLSVDSKGANSKPTSERIPDTDAPPTPPSAISSPIQFTRKQLALKILGLKVAASLHWNLDTLELNLAPQIQHQLMQDLVYMATDAGFAVPPQEIPAELMQKPQVIFALTLYHRWTLRFPVKAALYAKSSKIYMHMPGMQTDGGYPPLNQNFEKILRMCEGNRTHSIKFLEEVLSMYELTSNSSRRETKKIKVPPMEAFIHLTEDSDEVIHNWDAGDILITQFELAMQIHFDLCYNYFFYGQHDLAKKHILGCRENSNLLEREVSIYGYGPHKVVPWGEFYYASMTKDDIIGYIRALNLGQEVLNEEASLLQKLQESIANHYTGIIAVLQADNLAREIPMIHRQVVELDIQGSASSGAFTVARDLLNRVSALNAVRYALEGGIPSTHPDFLNKFKTVGIKFFDLLFWALGPVLLSDLSLEDWQHLRMFFLHLITSQCRLPIDRIDEYMKKHIGEAAGNIRKKLITDEQLSIILNDPINNDDENMEIPRELLTDDWETPDFDFKTVPELDMGRLKKRLIEASTADDVRMCLVKLAMMSPSLPLWKLSPSWKPAGVLGNALMSLNRGFLQDFGYVVSGAARARAESGSARTALSLLSVLEGEARSQLGGGTDPVLYRLCRQLSWEVLLLQVNVMLSEWPHHRLNLTALANKCKACIAAANSGDGIIPRPQVLEACWSCLVNSCEWEGLGVTTGPGELASALCAACFELQRGKGSRKFPRPLWDYALSVYSNGPNISMKRSAGGMSHAREMMNAAAESRNAFNGFLATLREPLAISIMMSLLARIHNLLIDDNSLELNTEYTSLWPSSVSNINNYNIKFVLESLTDMLERSLKLYPYNTSW
Summary
Description
Component of the Integrator complex, a complex involved in the transcription of small nuclear RNAs (snRNA) and their 3'-box-dependent processing (PubMed:21078872, PubMed:23097424). Involved in the 3'-end processing of the U7 snRNA, and also the spliceosomal snRNAs U1, U2, U4 and U5 (PubMed:21078872, PubMed:23097424).
Subunit
Belongs to the multiprotein complex Integrator, at least composed of IntS1, IntS2, IntS3, IntS4, omd/IntS5, IntS6, defl/IntS7, IntS8, IntS9, IntS10, IntS11, IntS12, asun/IntS13 and IntS14.
Similarity
Belongs to the dwarfin/SMAD family.
Belongs to the Integrator subunit 8 family.
Belongs to the Integrator subunit 8 family.
Keywords
Complete proteome
Nucleus
Reference proteome
Feature
chain Integrator complex subunit 8
Uniprot
H9JE59
A0A437B537
A0A2A4JR35
A0A212F005
A0A2H1W3B4
A0A194PKN4
+ More
A0A194R7P8 A0A0L7KT01 A0A0L7RH99 A0A195B7C7 A0A158NGY0 A0A195DWH2 E2AIH7 F4X124 A0A151XD29 A0A195FQ30 E2BWB0 A0A026X0Q8 A0A087ZZ34 A0A0M9A3Y3 A0A2J7PL41 A0A154PB00 A0A2A3EF35 A0A151ILM0 K7J228 A0A067RNU6 A0A182GK56 A0A232F5G6 A0A310SJD7 Q178M0 A0A1S4FC20 A0A0C9QTU2 A0A1Q3G202 W5JPK3 A0A182YDD3 A0A182XCB1 A0A1S4H7K5 A0A182TSX6 A0A182LPJ5 A0A182QPP1 A0A182P9D6 A0A182UYZ3 A0A2P8XUV1 A0A182WF50 A0A182MP25 A0A1A9WIS6 Q7Q324 A0A1Y1MIF8 D6WSC4 A0A182RFB3 E0VUJ0 A0A182JBU6 A0A1I8PH80 A0A0L0CQW0 A0A1B0B7E5 A0A1A9ZGI3 A0A1A9XP18 A0A0P5NCZ6 A0A0P5WNF3 A0A0P5SPL8 A0A1I8MH08 A0A182K240 A0A034VK54 B4QIG5 A0A1B0GI48 A1ZAK1 B4HTA0 B3NP62 A0A0A1WDZ2 A0A1Y1MIK6 A0A1W4UL65 A0A0P4XDT3 A0A084WBF4 W8BRV2 A0A182MXX2 B4P5V0 A0A0P5FX38 A0A1B6FX52 A0A182FTC9 A0A182HNL2 A0A3B0JU79 A0A0N7ZPS8 B4LM91 A0A1B0GNW5 B3MDA4 B4GIM2 B5DUR9 A0A1B6JHI1 B4MR83 B4KNM0 A0A0P5X4E1 A0A146LFG5 A0A0K8TK26 A0A0A9YL24 A0A0P5HG84 A0A0A9YMJ4
A0A194R7P8 A0A0L7KT01 A0A0L7RH99 A0A195B7C7 A0A158NGY0 A0A195DWH2 E2AIH7 F4X124 A0A151XD29 A0A195FQ30 E2BWB0 A0A026X0Q8 A0A087ZZ34 A0A0M9A3Y3 A0A2J7PL41 A0A154PB00 A0A2A3EF35 A0A151ILM0 K7J228 A0A067RNU6 A0A182GK56 A0A232F5G6 A0A310SJD7 Q178M0 A0A1S4FC20 A0A0C9QTU2 A0A1Q3G202 W5JPK3 A0A182YDD3 A0A182XCB1 A0A1S4H7K5 A0A182TSX6 A0A182LPJ5 A0A182QPP1 A0A182P9D6 A0A182UYZ3 A0A2P8XUV1 A0A182WF50 A0A182MP25 A0A1A9WIS6 Q7Q324 A0A1Y1MIF8 D6WSC4 A0A182RFB3 E0VUJ0 A0A182JBU6 A0A1I8PH80 A0A0L0CQW0 A0A1B0B7E5 A0A1A9ZGI3 A0A1A9XP18 A0A0P5NCZ6 A0A0P5WNF3 A0A0P5SPL8 A0A1I8MH08 A0A182K240 A0A034VK54 B4QIG5 A0A1B0GI48 A1ZAK1 B4HTA0 B3NP62 A0A0A1WDZ2 A0A1Y1MIK6 A0A1W4UL65 A0A0P4XDT3 A0A084WBF4 W8BRV2 A0A182MXX2 B4P5V0 A0A0P5FX38 A0A1B6FX52 A0A182FTC9 A0A182HNL2 A0A3B0JU79 A0A0N7ZPS8 B4LM91 A0A1B0GNW5 B3MDA4 B4GIM2 B5DUR9 A0A1B6JHI1 B4MR83 B4KNM0 A0A0P5X4E1 A0A146LFG5 A0A0K8TK26 A0A0A9YL24 A0A0P5HG84 A0A0A9YMJ4
Pubmed
19121390
22118469
26354079
26227816
21347285
20798317
+ More
21719571 24508170 30249741 20075255 24845553 26483478 28648823 17510324 20920257 23761445 25244985 12364791 20966253 29403074 28004739 18362917 19820115 20566863 26108605 25315136 25348373 17994087 22936249 10731132 12537572 10731137 12537569 21078872 23097424 25830018 24438588 24495485 17550304 15632085 26823975 25401762
21719571 24508170 30249741 20075255 24845553 26483478 28648823 17510324 20920257 23761445 25244985 12364791 20966253 29403074 28004739 18362917 19820115 20566863 26108605 25315136 25348373 17994087 22936249 10731132 12537572 10731137 12537569 21078872 23097424 25830018 24438588 24495485 17550304 15632085 26823975 25401762
EMBL
BABH01002625
BABH01002626
RSAL01000157
RVE45592.1
NWSH01000840
PCG73913.1
+ More
AGBW02011197 OWR47070.1 ODYU01005917 SOQ47312.1 KQ459602 KPI93294.1 KQ460615 KPJ13539.1 JTDY01006242 KOB66176.1 KQ414596 KOC70096.1 KQ976574 KYM80157.1 ADTU01015319 KQ980204 KYN17238.1 GL439791 EFN66767.1 GL888525 EGI59835.1 KQ982294 KYQ58284.1 KQ981387 KYN42029.1 GL451103 EFN80014.1 KK107063 QOIP01000005 EZA60979.1 RLU22451.1 KQ435765 KOX75369.1 NEVH01024527 PNF17053.1 KQ434864 KZC09011.1 KZ288269 PBC29892.1 KQ977104 KYN05764.1 KK852513 KDR22280.1 JXUM01069301 KQ562544 KXJ75658.1 NNAY01000882 OXU26076.1 KQ759820 OAD62741.1 CH477362 EAT42622.1 GBYB01007159 JAG76926.1 GFDL01001198 JAV33847.1 ADMH02000540 ETN66056.1 AAAB01008966 AXCN02002114 PYGN01001311 PSN35787.1 AXCM01000772 EAA13028.4 GEZM01030562 JAV85494.1 KQ971352 EFA06620.1 DS235786 EEB17046.1 JRES01000049 KNC34577.1 JXJN01009571 GDIQ01147168 JAL04558.1 GDIP01083880 LRGB01003024 JAM19835.1 KZS05013.1 GDIP01140484 JAL63230.1 GAKP01016083 JAC42869.1 CM000362 CM002911 EDX07412.1 KMY94376.1 AJWK01011291 AE013599 BT044437 AL031863 AY058406 AAL13635.1 ACH92502.1 CAA21313.1 CH480816 EDW48201.1 CH954179 EDV55701.1 GBXI01017406 JAC96885.1 GEZM01030565 JAV85493.1 GDIP01244556 JAI78845.1 ATLV01022351 KE525331 KFB47548.1 GAMC01010574 JAB95981.1 CM000158 EDW91866.1 GDIQ01256268 JAJ95456.1 GECZ01014993 JAS54776.1 APCN01001942 APCN01001943 OUUW01000001 SPP74628.1 GDIP01224513 JAI98888.1 CH940648 EDW60969.1 AJVK01031073 AJVK01031074 AJVK01031075 CH902619 EDV37437.1 CH479183 EDW36342.1 CH674335 EDY71638.1 GECU01009353 JAS98353.1 CH963849 EDW74622.1 CH933808 EDW10005.1 GDIP01078026 JAM25689.1 GDHC01011616 JAQ07013.1 GBRD01000101 JAG65720.1 GBHO01011283 JAG32321.1 GDIQ01227273 JAK24452.1 GBHO01011281 JAG32323.1
AGBW02011197 OWR47070.1 ODYU01005917 SOQ47312.1 KQ459602 KPI93294.1 KQ460615 KPJ13539.1 JTDY01006242 KOB66176.1 KQ414596 KOC70096.1 KQ976574 KYM80157.1 ADTU01015319 KQ980204 KYN17238.1 GL439791 EFN66767.1 GL888525 EGI59835.1 KQ982294 KYQ58284.1 KQ981387 KYN42029.1 GL451103 EFN80014.1 KK107063 QOIP01000005 EZA60979.1 RLU22451.1 KQ435765 KOX75369.1 NEVH01024527 PNF17053.1 KQ434864 KZC09011.1 KZ288269 PBC29892.1 KQ977104 KYN05764.1 KK852513 KDR22280.1 JXUM01069301 KQ562544 KXJ75658.1 NNAY01000882 OXU26076.1 KQ759820 OAD62741.1 CH477362 EAT42622.1 GBYB01007159 JAG76926.1 GFDL01001198 JAV33847.1 ADMH02000540 ETN66056.1 AAAB01008966 AXCN02002114 PYGN01001311 PSN35787.1 AXCM01000772 EAA13028.4 GEZM01030562 JAV85494.1 KQ971352 EFA06620.1 DS235786 EEB17046.1 JRES01000049 KNC34577.1 JXJN01009571 GDIQ01147168 JAL04558.1 GDIP01083880 LRGB01003024 JAM19835.1 KZS05013.1 GDIP01140484 JAL63230.1 GAKP01016083 JAC42869.1 CM000362 CM002911 EDX07412.1 KMY94376.1 AJWK01011291 AE013599 BT044437 AL031863 AY058406 AAL13635.1 ACH92502.1 CAA21313.1 CH480816 EDW48201.1 CH954179 EDV55701.1 GBXI01017406 JAC96885.1 GEZM01030565 JAV85493.1 GDIP01244556 JAI78845.1 ATLV01022351 KE525331 KFB47548.1 GAMC01010574 JAB95981.1 CM000158 EDW91866.1 GDIQ01256268 JAJ95456.1 GECZ01014993 JAS54776.1 APCN01001942 APCN01001943 OUUW01000001 SPP74628.1 GDIP01224513 JAI98888.1 CH940648 EDW60969.1 AJVK01031073 AJVK01031074 AJVK01031075 CH902619 EDV37437.1 CH479183 EDW36342.1 CH674335 EDY71638.1 GECU01009353 JAS98353.1 CH963849 EDW74622.1 CH933808 EDW10005.1 GDIP01078026 JAM25689.1 GDHC01011616 JAQ07013.1 GBRD01000101 JAG65720.1 GBHO01011283 JAG32321.1 GDIQ01227273 JAK24452.1 GBHO01011281 JAG32323.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053268
UP000053240
+ More
UP000037510 UP000053825 UP000078540 UP000005205 UP000078492 UP000000311 UP000007755 UP000075809 UP000078541 UP000008237 UP000053097 UP000279307 UP000005203 UP000053105 UP000235965 UP000076502 UP000242457 UP000078542 UP000002358 UP000027135 UP000069940 UP000249989 UP000215335 UP000008820 UP000000673 UP000076408 UP000076407 UP000075902 UP000075882 UP000075886 UP000075885 UP000075903 UP000245037 UP000075920 UP000075883 UP000091820 UP000007062 UP000007266 UP000075900 UP000009046 UP000075880 UP000095300 UP000037069 UP000092460 UP000092445 UP000092443 UP000076858 UP000095301 UP000075881 UP000000304 UP000092461 UP000000803 UP000001292 UP000008711 UP000192221 UP000030765 UP000075884 UP000002282 UP000069272 UP000075840 UP000268350 UP000008792 UP000092462 UP000007801 UP000008744 UP000001819 UP000007798 UP000009192
UP000037510 UP000053825 UP000078540 UP000005205 UP000078492 UP000000311 UP000007755 UP000075809 UP000078541 UP000008237 UP000053097 UP000279307 UP000005203 UP000053105 UP000235965 UP000076502 UP000242457 UP000078542 UP000002358 UP000027135 UP000069940 UP000249989 UP000215335 UP000008820 UP000000673 UP000076408 UP000076407 UP000075902 UP000075882 UP000075886 UP000075885 UP000075903 UP000245037 UP000075920 UP000075883 UP000091820 UP000007062 UP000007266 UP000075900 UP000009046 UP000075880 UP000095300 UP000037069 UP000092460 UP000092445 UP000092443 UP000076858 UP000095301 UP000075881 UP000000304 UP000092461 UP000000803 UP000001292 UP000008711 UP000192221 UP000030765 UP000075884 UP000002282 UP000069272 UP000075840 UP000268350 UP000008792 UP000092462 UP000007801 UP000008744 UP000001819 UP000007798 UP000009192
Pfam
Interpro
IPR038751
Int8
+ More
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR003619 MAD_homology1_Dwarfin-type
IPR036578 SMAD_MH1_sf
IPR008984 SMAD_FHA_dom_sf
IPR013790 Dwarfin
IPR017855 SMAD-like_dom_sf
IPR001132 SMAD_dom_Dwarfin-type
IPR013019 MAD_homology_MH1
IPR014784 Cu2_ascorb_mOase-like_C
IPR005018 DOMON_domain
IPR036939 Cu2_ascorb_mOase_N_sf
IPR008977 PHM/PNGase_F_dom_sf
IPR024548 Cu2_monoox_C
IPR028460 Tbh/DBH
IPR020611 Cu2_ascorb_mOase_CS-1
IPR000323 Cu2_ascorb_mOase_N
IPR027417 P-loop_NTPase
IPR000629 RNA-helicase_DEAD-box_CS
IPR014001 Helicase_ATP-bd
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR003619 MAD_homology1_Dwarfin-type
IPR036578 SMAD_MH1_sf
IPR008984 SMAD_FHA_dom_sf
IPR013790 Dwarfin
IPR017855 SMAD-like_dom_sf
IPR001132 SMAD_dom_Dwarfin-type
IPR013019 MAD_homology_MH1
IPR014784 Cu2_ascorb_mOase-like_C
IPR005018 DOMON_domain
IPR036939 Cu2_ascorb_mOase_N_sf
IPR008977 PHM/PNGase_F_dom_sf
IPR024548 Cu2_monoox_C
IPR028460 Tbh/DBH
IPR020611 Cu2_ascorb_mOase_CS-1
IPR000323 Cu2_ascorb_mOase_N
IPR027417 P-loop_NTPase
IPR000629 RNA-helicase_DEAD-box_CS
IPR014001 Helicase_ATP-bd
IPR014014 RNA_helicase_DEAD_Q_motif
IPR011545 DEAD/DEAH_box_helicase_dom
SUPFAM
ProteinModelPortal
H9JE59
A0A437B537
A0A2A4JR35
A0A212F005
A0A2H1W3B4
A0A194PKN4
+ More
A0A194R7P8 A0A0L7KT01 A0A0L7RH99 A0A195B7C7 A0A158NGY0 A0A195DWH2 E2AIH7 F4X124 A0A151XD29 A0A195FQ30 E2BWB0 A0A026X0Q8 A0A087ZZ34 A0A0M9A3Y3 A0A2J7PL41 A0A154PB00 A0A2A3EF35 A0A151ILM0 K7J228 A0A067RNU6 A0A182GK56 A0A232F5G6 A0A310SJD7 Q178M0 A0A1S4FC20 A0A0C9QTU2 A0A1Q3G202 W5JPK3 A0A182YDD3 A0A182XCB1 A0A1S4H7K5 A0A182TSX6 A0A182LPJ5 A0A182QPP1 A0A182P9D6 A0A182UYZ3 A0A2P8XUV1 A0A182WF50 A0A182MP25 A0A1A9WIS6 Q7Q324 A0A1Y1MIF8 D6WSC4 A0A182RFB3 E0VUJ0 A0A182JBU6 A0A1I8PH80 A0A0L0CQW0 A0A1B0B7E5 A0A1A9ZGI3 A0A1A9XP18 A0A0P5NCZ6 A0A0P5WNF3 A0A0P5SPL8 A0A1I8MH08 A0A182K240 A0A034VK54 B4QIG5 A0A1B0GI48 A1ZAK1 B4HTA0 B3NP62 A0A0A1WDZ2 A0A1Y1MIK6 A0A1W4UL65 A0A0P4XDT3 A0A084WBF4 W8BRV2 A0A182MXX2 B4P5V0 A0A0P5FX38 A0A1B6FX52 A0A182FTC9 A0A182HNL2 A0A3B0JU79 A0A0N7ZPS8 B4LM91 A0A1B0GNW5 B3MDA4 B4GIM2 B5DUR9 A0A1B6JHI1 B4MR83 B4KNM0 A0A0P5X4E1 A0A146LFG5 A0A0K8TK26 A0A0A9YL24 A0A0P5HG84 A0A0A9YMJ4
A0A194R7P8 A0A0L7KT01 A0A0L7RH99 A0A195B7C7 A0A158NGY0 A0A195DWH2 E2AIH7 F4X124 A0A151XD29 A0A195FQ30 E2BWB0 A0A026X0Q8 A0A087ZZ34 A0A0M9A3Y3 A0A2J7PL41 A0A154PB00 A0A2A3EF35 A0A151ILM0 K7J228 A0A067RNU6 A0A182GK56 A0A232F5G6 A0A310SJD7 Q178M0 A0A1S4FC20 A0A0C9QTU2 A0A1Q3G202 W5JPK3 A0A182YDD3 A0A182XCB1 A0A1S4H7K5 A0A182TSX6 A0A182LPJ5 A0A182QPP1 A0A182P9D6 A0A182UYZ3 A0A2P8XUV1 A0A182WF50 A0A182MP25 A0A1A9WIS6 Q7Q324 A0A1Y1MIF8 D6WSC4 A0A182RFB3 E0VUJ0 A0A182JBU6 A0A1I8PH80 A0A0L0CQW0 A0A1B0B7E5 A0A1A9ZGI3 A0A1A9XP18 A0A0P5NCZ6 A0A0P5WNF3 A0A0P5SPL8 A0A1I8MH08 A0A182K240 A0A034VK54 B4QIG5 A0A1B0GI48 A1ZAK1 B4HTA0 B3NP62 A0A0A1WDZ2 A0A1Y1MIK6 A0A1W4UL65 A0A0P4XDT3 A0A084WBF4 W8BRV2 A0A182MXX2 B4P5V0 A0A0P5FX38 A0A1B6FX52 A0A182FTC9 A0A182HNL2 A0A3B0JU79 A0A0N7ZPS8 B4LM91 A0A1B0GNW5 B3MDA4 B4GIM2 B5DUR9 A0A1B6JHI1 B4MR83 B4KNM0 A0A0P5X4E1 A0A146LFG5 A0A0K8TK26 A0A0A9YL24 A0A0P5HG84 A0A0A9YMJ4
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
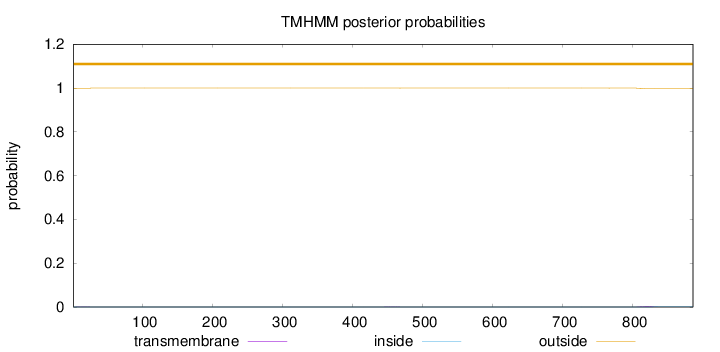
Length:
886
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06668
Exp number, first 60 AAs:
0.00544
Total prob of N-in:
0.00056
outside
1 - 886
Population Genetic Test Statistics
Pi
233.82538
Theta
191.674114
Tajima's D
0.991235
CLR
0.001161
CSRT
0.653717314134293
Interpretation
Uncertain
