Gene
KWMTBOMO08933
Annotation
PREDICTED:_uncharacterized_protein_LOC101737114_isoform_X2_[Bombyx_mori]
Transcription factor
Location in the cell
Extracellular Reliability : 2.613
Sequence
CDS
ATGGAAAACCATGGGAAAGGTGGACCTTTGGCCACAACGGTGTATCAAGTTCCTGCGATTGAGAAGCCGTTATTGGGTCCGGGGAACACAGACACAATTTGCCCGTTTTGTCGAGCGTCCATTAGGAGTGCCATAAAGTACAGTATTACCTCGAAAACGCACATTGCCGCTGCTCTCTGGTGCTTGGTTTGTTGCCTCTGTTGCATGCCGTATGCGTCGGATACATGTAAAAACGTGGACCATTTTTGTCCAAATTGCCATAAATACATCGGAACTTACGCGAGGTAA
Protein
MENHGKGGPLATTVYQVPAIEKPLLGPGNTDTICPFCRASIRSAIKYSITSKTHIAAALWCLVCCLCCMPYASDTCKNVDHFCPNCHKYIGTYAR
Summary
Uniprot
A0A437B516
A0A2H1W2R2
A0A2A4JQ73
A0A1Q3FMI6
A0A212FFW5
B0X8G3
+ More
A0A1L8E611 A0A1B0CTZ3 T1DFW1 A0A034UX67 A0A023EE14 A0A023EBS0 A0A212FFW2 A0A1L8EIY1 A0A182GJR8 A0A1L8E331 A0A1I8NQQ1 A0A1L8E2U5 A0A0K8U1X9 A0A1L8E637 A0A0A1XAV4 A0A1B0GQM0 A0A2M4CXL3 A0A2M4CXZ0 A0A182G7S7 A0A1S4FG64 Q172M2 A0A2W1BQA8 A0A2H1WAD5 A0A2W1BYV4 A0A437BIX7 A0A182PVK7 A7UV78 A0A437BJ34 A0A2M4C4E3 A0A2M4AS09 A8CA04 A0A2W1BNA3 A0A1B0DR17 W5JPJ7 A0A182F3N1 T1EAL8 A0A182VSL0 A0A2W1BM26 A0A182NAP6 A0A182YMU5 Q172M0 A0A182V1S5 A0A182RWG6 A0A182X867 A0A1S4H0Q8 A0A1C7CZ26 A0A182HZ05 A0A2M4AXT4 A0A2M4C324 A0A336N1I9 A0A182VL92 W8CB75 A0A182TGD0 A0A182KYN3 A7UV77 A0A182HZ06 A0A1L8E2T6 A0A2A4JVU8 A0A1Q3FKN6 U5ES92 A0A1L8E397 A0A182QYK8 B0X8G2 A0A182X868 S4P885 A0A084VYA4 A0A1B0CVX8 A0A2W1BVG7 A0A023EH32 A0A182M0D7 A0A2A4JWY3 A0A182GCI1 A0A182JDM5 A0A182F3N0 A0A182QDH4 A0A182VSK9 A0A182RWG5 A0A1J1INJ0 A0A2W1BN81 A0A1S4FG68 A0A161MIM9 A0A194RW35 A0A2A4JBS3 A0A182TNI9 H9IUF6 A0A2A4JW93 A0A182YMU6 A0A182PVK6 A0A0K8TL77 A0A2W1BW29 A0A084VYA2 A0A1B6EBM5 Q172M1
A0A1L8E611 A0A1B0CTZ3 T1DFW1 A0A034UX67 A0A023EE14 A0A023EBS0 A0A212FFW2 A0A1L8EIY1 A0A182GJR8 A0A1L8E331 A0A1I8NQQ1 A0A1L8E2U5 A0A0K8U1X9 A0A1L8E637 A0A0A1XAV4 A0A1B0GQM0 A0A2M4CXL3 A0A2M4CXZ0 A0A182G7S7 A0A1S4FG64 Q172M2 A0A2W1BQA8 A0A2H1WAD5 A0A2W1BYV4 A0A437BIX7 A0A182PVK7 A7UV78 A0A437BJ34 A0A2M4C4E3 A0A2M4AS09 A8CA04 A0A2W1BNA3 A0A1B0DR17 W5JPJ7 A0A182F3N1 T1EAL8 A0A182VSL0 A0A2W1BM26 A0A182NAP6 A0A182YMU5 Q172M0 A0A182V1S5 A0A182RWG6 A0A182X867 A0A1S4H0Q8 A0A1C7CZ26 A0A182HZ05 A0A2M4AXT4 A0A2M4C324 A0A336N1I9 A0A182VL92 W8CB75 A0A182TGD0 A0A182KYN3 A7UV77 A0A182HZ06 A0A1L8E2T6 A0A2A4JVU8 A0A1Q3FKN6 U5ES92 A0A1L8E397 A0A182QYK8 B0X8G2 A0A182X868 S4P885 A0A084VYA4 A0A1B0CVX8 A0A2W1BVG7 A0A023EH32 A0A182M0D7 A0A2A4JWY3 A0A182GCI1 A0A182JDM5 A0A182F3N0 A0A182QDH4 A0A182VSK9 A0A182RWG5 A0A1J1INJ0 A0A2W1BN81 A0A1S4FG68 A0A161MIM9 A0A194RW35 A0A2A4JBS3 A0A182TNI9 H9IUF6 A0A2A4JW93 A0A182YMU6 A0A182PVK6 A0A0K8TL77 A0A2W1BW29 A0A084VYA2 A0A1B6EBM5 Q172M1
Pubmed
EMBL
RSAL01000157
RVE45591.1
ODYU01005917
SOQ47313.1
NWSH01000840
PCG73916.1
+ More
GFDL01006278 JAV28767.1 AGBW02008757 OWR52624.1 DS232488 EDS42499.1 GFDG01004683 JAV14116.1 AJWK01028209 GALA01000412 JAA94440.1 GAKP01023392 JAC35566.1 GAPW01006739 JAC06859.1 GAPW01006670 JAC06928.1 OWR52625.1 GFDG01000195 JAV18604.1 JXUM01068757 JXUM01068758 JXUM01068759 JXUM01068760 JXUM01068761 JXUM01068762 JXUM01093175 KQ564039 KQ562516 KXJ72882.1 KXJ75727.1 GFDF01001040 JAV13044.1 GFDF01001021 JAV13063.1 GDHF01031984 JAI20330.1 GFDG01004685 JAV14114.1 GBXI01005868 JAD08424.1 AJVK01017302 GGFL01005894 MBW70072.1 GGFL01005893 MBW70071.1 JXUM01046825 JXUM01054329 KQ561816 KQ561492 KXJ77441.1 KXJ78397.1 CH477434 EAT40986.1 KZ149983 PZC75755.1 ODYU01007335 SOQ50013.1 KZ149918 PZC77856.1 RSAL01000049 RVE50382.1 AAAB01008984 EDO63390.1 RVE50383.1 GGFJ01011039 MBW60180.1 GGFK01010240 MBW43561.1 EU035828 ABV44730.1 PZC75751.1 AJVK01019546 AJVK01019547 ADMH02000911 ETN64689.1 GAMD01000923 JAB00668.1 KZ150055 PZC74327.1 EAT40988.1 APCN01005837 GGFK01012234 MBW45555.1 GGFJ01010267 MBW59408.1 UFQT01002583 UFQT01003106 SSX33713.1 SSX34587.1 GAMC01000699 JAC05857.1 EDO63389.1 GFDF01001031 JAV13053.1 NWSH01000518 PCG75906.1 GFDL01006896 JAV28149.1 GANO01003353 JAB56518.1 GFDF01001032 JAV13052.1 AXCN02000669 EDS42498.1 GAIX01006056 JAA86504.1 ATLV01018349 KE525231 KFB42948.1 AJWK01031388 PZC75753.1 GAPW01005437 JAC08161.1 AXCM01014984 PCG75902.1 JXUM01054326 JXUM01054327 JXUM01054328 KXJ77440.1 AXCN02000670 CVRI01000055 CRL01124.1 PZC74326.1 GEMB01006008 JAR97317.1 KQ459700 KPJ20401.1 NWSH01002225 PCG68882.1 BABH01001130 PCG75904.1 GDAI01002938 JAI14665.1 PZC77855.1 ATLV01018347 KFB42946.1 GEDC01001960 JAS35338.1 EAT40987.1
GFDL01006278 JAV28767.1 AGBW02008757 OWR52624.1 DS232488 EDS42499.1 GFDG01004683 JAV14116.1 AJWK01028209 GALA01000412 JAA94440.1 GAKP01023392 JAC35566.1 GAPW01006739 JAC06859.1 GAPW01006670 JAC06928.1 OWR52625.1 GFDG01000195 JAV18604.1 JXUM01068757 JXUM01068758 JXUM01068759 JXUM01068760 JXUM01068761 JXUM01068762 JXUM01093175 KQ564039 KQ562516 KXJ72882.1 KXJ75727.1 GFDF01001040 JAV13044.1 GFDF01001021 JAV13063.1 GDHF01031984 JAI20330.1 GFDG01004685 JAV14114.1 GBXI01005868 JAD08424.1 AJVK01017302 GGFL01005894 MBW70072.1 GGFL01005893 MBW70071.1 JXUM01046825 JXUM01054329 KQ561816 KQ561492 KXJ77441.1 KXJ78397.1 CH477434 EAT40986.1 KZ149983 PZC75755.1 ODYU01007335 SOQ50013.1 KZ149918 PZC77856.1 RSAL01000049 RVE50382.1 AAAB01008984 EDO63390.1 RVE50383.1 GGFJ01011039 MBW60180.1 GGFK01010240 MBW43561.1 EU035828 ABV44730.1 PZC75751.1 AJVK01019546 AJVK01019547 ADMH02000911 ETN64689.1 GAMD01000923 JAB00668.1 KZ150055 PZC74327.1 EAT40988.1 APCN01005837 GGFK01012234 MBW45555.1 GGFJ01010267 MBW59408.1 UFQT01002583 UFQT01003106 SSX33713.1 SSX34587.1 GAMC01000699 JAC05857.1 EDO63389.1 GFDF01001031 JAV13053.1 NWSH01000518 PCG75906.1 GFDL01006896 JAV28149.1 GANO01003353 JAB56518.1 GFDF01001032 JAV13052.1 AXCN02000669 EDS42498.1 GAIX01006056 JAA86504.1 ATLV01018349 KE525231 KFB42948.1 AJWK01031388 PZC75753.1 GAPW01005437 JAC08161.1 AXCM01014984 PCG75902.1 JXUM01054326 JXUM01054327 JXUM01054328 KXJ77440.1 AXCN02000670 CVRI01000055 CRL01124.1 PZC74326.1 GEMB01006008 JAR97317.1 KQ459700 KPJ20401.1 NWSH01002225 PCG68882.1 BABH01001130 PCG75904.1 GDAI01002938 JAI14665.1 PZC77855.1 ATLV01018347 KFB42946.1 GEDC01001960 JAS35338.1 EAT40987.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000002320
UP000092461
UP000069940
+ More
UP000249989 UP000095300 UP000092462 UP000008820 UP000075885 UP000007062 UP000000673 UP000069272 UP000075920 UP000075884 UP000076408 UP000075903 UP000075900 UP000076407 UP000075882 UP000075840 UP000075902 UP000075886 UP000030765 UP000075883 UP000075880 UP000183832 UP000053240 UP000005204
UP000249989 UP000095300 UP000092462 UP000008820 UP000075885 UP000007062 UP000000673 UP000069272 UP000075920 UP000075884 UP000076408 UP000075903 UP000075900 UP000076407 UP000075882 UP000075840 UP000075902 UP000075886 UP000030765 UP000075883 UP000075880 UP000183832 UP000053240 UP000005204
PRIDE
Pfam
PF10601 zf-LITAF-like
ProteinModelPortal
A0A437B516
A0A2H1W2R2
A0A2A4JQ73
A0A1Q3FMI6
A0A212FFW5
B0X8G3
+ More
A0A1L8E611 A0A1B0CTZ3 T1DFW1 A0A034UX67 A0A023EE14 A0A023EBS0 A0A212FFW2 A0A1L8EIY1 A0A182GJR8 A0A1L8E331 A0A1I8NQQ1 A0A1L8E2U5 A0A0K8U1X9 A0A1L8E637 A0A0A1XAV4 A0A1B0GQM0 A0A2M4CXL3 A0A2M4CXZ0 A0A182G7S7 A0A1S4FG64 Q172M2 A0A2W1BQA8 A0A2H1WAD5 A0A2W1BYV4 A0A437BIX7 A0A182PVK7 A7UV78 A0A437BJ34 A0A2M4C4E3 A0A2M4AS09 A8CA04 A0A2W1BNA3 A0A1B0DR17 W5JPJ7 A0A182F3N1 T1EAL8 A0A182VSL0 A0A2W1BM26 A0A182NAP6 A0A182YMU5 Q172M0 A0A182V1S5 A0A182RWG6 A0A182X867 A0A1S4H0Q8 A0A1C7CZ26 A0A182HZ05 A0A2M4AXT4 A0A2M4C324 A0A336N1I9 A0A182VL92 W8CB75 A0A182TGD0 A0A182KYN3 A7UV77 A0A182HZ06 A0A1L8E2T6 A0A2A4JVU8 A0A1Q3FKN6 U5ES92 A0A1L8E397 A0A182QYK8 B0X8G2 A0A182X868 S4P885 A0A084VYA4 A0A1B0CVX8 A0A2W1BVG7 A0A023EH32 A0A182M0D7 A0A2A4JWY3 A0A182GCI1 A0A182JDM5 A0A182F3N0 A0A182QDH4 A0A182VSK9 A0A182RWG5 A0A1J1INJ0 A0A2W1BN81 A0A1S4FG68 A0A161MIM9 A0A194RW35 A0A2A4JBS3 A0A182TNI9 H9IUF6 A0A2A4JW93 A0A182YMU6 A0A182PVK6 A0A0K8TL77 A0A2W1BW29 A0A084VYA2 A0A1B6EBM5 Q172M1
A0A1L8E611 A0A1B0CTZ3 T1DFW1 A0A034UX67 A0A023EE14 A0A023EBS0 A0A212FFW2 A0A1L8EIY1 A0A182GJR8 A0A1L8E331 A0A1I8NQQ1 A0A1L8E2U5 A0A0K8U1X9 A0A1L8E637 A0A0A1XAV4 A0A1B0GQM0 A0A2M4CXL3 A0A2M4CXZ0 A0A182G7S7 A0A1S4FG64 Q172M2 A0A2W1BQA8 A0A2H1WAD5 A0A2W1BYV4 A0A437BIX7 A0A182PVK7 A7UV78 A0A437BJ34 A0A2M4C4E3 A0A2M4AS09 A8CA04 A0A2W1BNA3 A0A1B0DR17 W5JPJ7 A0A182F3N1 T1EAL8 A0A182VSL0 A0A2W1BM26 A0A182NAP6 A0A182YMU5 Q172M0 A0A182V1S5 A0A182RWG6 A0A182X867 A0A1S4H0Q8 A0A1C7CZ26 A0A182HZ05 A0A2M4AXT4 A0A2M4C324 A0A336N1I9 A0A182VL92 W8CB75 A0A182TGD0 A0A182KYN3 A7UV77 A0A182HZ06 A0A1L8E2T6 A0A2A4JVU8 A0A1Q3FKN6 U5ES92 A0A1L8E397 A0A182QYK8 B0X8G2 A0A182X868 S4P885 A0A084VYA4 A0A1B0CVX8 A0A2W1BVG7 A0A023EH32 A0A182M0D7 A0A2A4JWY3 A0A182GCI1 A0A182JDM5 A0A182F3N0 A0A182QDH4 A0A182VSK9 A0A182RWG5 A0A1J1INJ0 A0A2W1BN81 A0A1S4FG68 A0A161MIM9 A0A194RW35 A0A2A4JBS3 A0A182TNI9 H9IUF6 A0A2A4JW93 A0A182YMU6 A0A182PVK6 A0A0K8TL77 A0A2W1BW29 A0A084VYA2 A0A1B6EBM5 Q172M1
Ontologies
GO
PANTHER
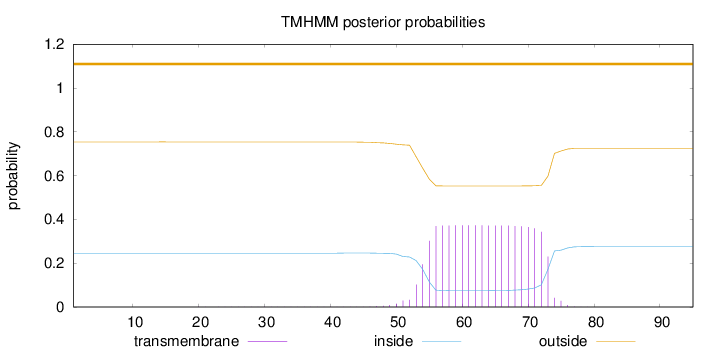
Topology
Length:
95
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.28129
Exp number, first 60 AAs:
2.5595
Total prob of N-in:
0.24603
outside
1 - 95
Population Genetic Test Statistics
Pi
226.50288
Theta
185.713883
Tajima's D
0.666356
CLR
0.441664
CSRT
0.566221688915554
Interpretation
Uncertain
