Gene
KWMTBOMO08931
Pre Gene Modal
BGIBMGA007807
Annotation
PREDICTED:_presenilins-associated_rhomboid-like_protein?_mitochondrial_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.812
Sequence
CDS
ATGTTTTCTAGAACTTTAAATGTTTCCGCAAATATTTGCAAACCACCACTATTCAATCAAGTTTGGCTTCCACCTAAGAACGGTCGATTGATCCGTAATTCATTTCACAATTCAAAAAGAGGTTCGCGGCAAGCTCTGAAACCTGACCCTTTGGAAAATCTACATATTGAAACTGGCCCGTTACATGCGAAAGGATTAGTGAAACCGCTTATTTTTACTGTGGGAGTCTCAGCAGCCAGTTTACTAGGCTGTGTAATATGGGAATATGAGAATCTACGGGTTCACGCTTCTTCACTACTACGAAGACCAGGAACGTGGCTGACTGCACAACAAAAAAAAAGTAAATCAGAGCCTGGTCCATTGAAAAAATGGTGGAACTCACTACGGGATAATGAGAAGGTGTTTTATCCAATTTTAGCTGCTAACTTACTAGTATTTGGAGCATGGCGCATTAGAGCTCTACAGCCTTTTATGGTCAAATACTTCTGTTCCAATCCTTCAGGAGGTGCAGTATGTCTTCCGATGGTTCTTTCAACATTCAGCCATTATTCACCATTGCACCTGGCTGCCAATATGTATGTTCTTTATAGTTTCATGCCTGCTGCTTTATCTTCAATGGGCAAAGAACAATTTACAGCCATGTATCTCAGTGCCGGTGTCATAAGCAGTTTTGCTAGTTTTCTTTACAAGGTTATGCTAAATCAACCAGGATTAAGCCTTGGAGCATCTGGTGCAATCATGTCAGTGCTGTCTTATGTGTGTATGCAATACCCCGACACAAAACTAAGCATCATATTTTTACCAATGTACACTTTTGCGGCTGGTTCTGCAATCAAAGTTATAATGGGAATAGATCTGGCTGGTGTTGTCTTGGGTTGGAAGTTCTTTGATCATGCTGCTCATCTTGGTGGAGCTATATTTGGAATTATATGGTGCCAATGGGGTTATTCTCACGTATGGGGTAACAGAGACAAATTCCTCCAGTATTATCACCACTTTAGAAAAAGCGATCCCGGAAGGTAG
Protein
MFSRTLNVSANICKPPLFNQVWLPPKNGRLIRNSFHNSKRGSRQALKPDPLENLHIETGPLHAKGLVKPLIFTVGVSAASLLGCVIWEYENLRVHASSLLRRPGTWLTAQQKKSKSEPGPLKKWWNSLRDNEKVFYPILAANLLVFGAWRIRALQPFMVKYFCSNPSGGAVCLPMVLSTFSHYSPLHLAANMYVLYSFMPAALSSMGKEQFTAMYLSAGVISSFASFLYKVMLNQPGLSLGASGAIMSVLSYVCMQYPDTKLSIIFLPMYTFAAGSAIKVIMGIDLAGVVLGWKFFDHAAHLGGAIFGIIWCQWGYSHVWGNRDKFLQYYHHFRKSDPGR
Summary
Similarity
Belongs to the synaptobrevin family.
Uniprot
A0A3S2LWI9
A0A194RCA1
A0A2A4JPJ0
A0A194PIZ5
I4DL50
A0A212ELH1
+ More
G3JWY3 A0A2P8XJE4 A0A1Y1MNY9 D6WL40 A0A182NRE1 A0A182PI63 A0A182QPL7 A0A182STB1 A0A182V8Q2 A0A182Y6L5 A0A182TQ58 A0A182HSG9 A0A182KCN9 A0A182LEH8 A0A182XAM8 Q7QCH5 A0A1Q3FEU1 A0A182RXQ3 A0A084W6L4 A0A0V0G6C3 Q16I97 A0A2J7Q205 A0A0P4VLT5 R4FLP2 A0A2M4ASR3 A0A2M4ASS3 B0X7U0 A0A2M4BSW3 A0A182W0T2 W5J9B7 A0A182FPB9 A0A182IL98 A0A336K0Q3 A0A182M9Q5 A0A1L8DTM3 A0A1W4WZ51 A0A2M4BS76 A0A1L8DTW0 U4UTS1 A0A067QYA2 A0A023ERX6 A0A182GQK3 A0A195C2S4 A0A0L0CM32 F4WD26 A0A151XDF7 A0A1B0DMJ2 A0A1I8N9M6 A0A1I8QA46 A0A158NID5 A0A1B6E5C0 A0A1A9WBA4 A0A1B0A7I5 A0A195BFG7 A0A1B0FQP0 B4MDW5 B4J3U7 A0A1A9UY58 A0A0Q9WAZ1 B4KSE3 A0A0A1XJQ2 B4MYM0 E0VBZ6 A0A0K8WF12 E1ZX17 A0A3L8DCW6 A0A2R7W342 A0A0R3NP36 Q28YK6 A0A034VGR2 G3WN22 A0A1I8QA10 W8C932 A0A146M7U5 B4HNW2 B4QC61 B4H8I2 A0A1W4UNP1 B3MG48 A1Z8R8 A0A3P8WEV8 Q8MQY3 A0A0M4E448 B4P5R1 A0A0P6CNX2 K9IXI8 A0A2D0STN9 A0A1J1IBW2 A0A1W4Z3M9 A0A151JNT4 B3NSA1
G3JWY3 A0A2P8XJE4 A0A1Y1MNY9 D6WL40 A0A182NRE1 A0A182PI63 A0A182QPL7 A0A182STB1 A0A182V8Q2 A0A182Y6L5 A0A182TQ58 A0A182HSG9 A0A182KCN9 A0A182LEH8 A0A182XAM8 Q7QCH5 A0A1Q3FEU1 A0A182RXQ3 A0A084W6L4 A0A0V0G6C3 Q16I97 A0A2J7Q205 A0A0P4VLT5 R4FLP2 A0A2M4ASR3 A0A2M4ASS3 B0X7U0 A0A2M4BSW3 A0A182W0T2 W5J9B7 A0A182FPB9 A0A182IL98 A0A336K0Q3 A0A182M9Q5 A0A1L8DTM3 A0A1W4WZ51 A0A2M4BS76 A0A1L8DTW0 U4UTS1 A0A067QYA2 A0A023ERX6 A0A182GQK3 A0A195C2S4 A0A0L0CM32 F4WD26 A0A151XDF7 A0A1B0DMJ2 A0A1I8N9M6 A0A1I8QA46 A0A158NID5 A0A1B6E5C0 A0A1A9WBA4 A0A1B0A7I5 A0A195BFG7 A0A1B0FQP0 B4MDW5 B4J3U7 A0A1A9UY58 A0A0Q9WAZ1 B4KSE3 A0A0A1XJQ2 B4MYM0 E0VBZ6 A0A0K8WF12 E1ZX17 A0A3L8DCW6 A0A2R7W342 A0A0R3NP36 Q28YK6 A0A034VGR2 G3WN22 A0A1I8QA10 W8C932 A0A146M7U5 B4HNW2 B4QC61 B4H8I2 A0A1W4UNP1 B3MG48 A1Z8R8 A0A3P8WEV8 Q8MQY3 A0A0M4E448 B4P5R1 A0A0P6CNX2 K9IXI8 A0A2D0STN9 A0A1J1IBW2 A0A1W4Z3M9 A0A151JNT4 B3NSA1
Pubmed
26354079
22651552
22118469
21840855
29403074
28004739
+ More
18362917 19820115 25244985 20966253 12364791 14747013 17210077 24438588 17510324 27129103 20920257 23761445 23537049 24845553 24945155 26483478 26108605 21719571 25315136 21347285 17994087 25830018 20566863 20798317 30249741 15632085 25348373 21709235 24495485 26823975 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24487278 17550304
18362917 19820115 25244985 20966253 12364791 14747013 17210077 24438588 17510324 27129103 20920257 23761445 23537049 24845553 24945155 26483478 26108605 21719571 25315136 21347285 17994087 25830018 20566863 20798317 30249741 15632085 25348373 21709235 24495485 26823975 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24487278 17550304
EMBL
RSAL01000157
RVE45586.1
KQ460615
KPJ13536.1
NWSH01000840
PCG73921.1
+ More
KQ459602 KPI93292.1 AK402018 BAM18640.1 AGBW02014057 OWR42336.1 JN030492 AEI27594.1 PYGN01001923 PSN32127.1 GEZM01025721 JAV87371.1 KQ971343 EFA03514.2 AXCN02001464 APCN01000278 AAAB01008859 EAA08059.4 GFDL01008971 JAV26074.1 ATLV01020849 KE525308 KFB45858.1 GECL01002884 JAP03240.1 CH478095 EAT33981.1 NEVH01019374 PNF22625.1 GDKW01003201 JAI53394.1 ACPB03015003 GAHY01002059 JAA75451.1 GGFK01010479 MBW43800.1 GGFK01010510 MBW43831.1 DS232463 EDS42159.1 GGFJ01006717 MBW55858.1 ADMH02002014 ETN59988.1 UFQS01000046 UFQT01000046 SSW98524.1 SSX18910.1 AXCM01000472 GFDF01004271 JAV09813.1 GGFJ01006721 MBW55862.1 GFDF01004270 JAV09814.1 KB632364 ERL93545.1 KK852880 KDR14418.1 GAPW01002137 JAC11461.1 JXUM01080774 KQ563206 KXJ74289.1 KQ978379 KYM94488.1 JRES01000204 KNC33302.1 GL888081 EGI67898.1 KQ982294 KYQ58395.1 AJVK01016832 ADTU01016577 GEDC01004190 JAS33108.1 KQ976491 KYM83321.1 CCAG010009454 CH940662 EDW58730.1 CH916367 EDW01530.1 KRF78436.1 CH933808 EDW08425.1 GBXI01003115 JAD11177.1 CH963894 EDW77209.2 DS235044 EEB10902.1 GDHF01002670 JAI49644.1 GL434950 EFN74267.1 QOIP01000010 RLU18171.1 KK854259 PTY13848.1 CM000071 KRT02699.1 EAL25960.2 GAKP01018239 JAC40713.1 AEFK01178545 AEFK01178546 AEFK01178547 GAMC01006996 JAB99559.1 GDHC01003989 JAQ14640.1 CH480816 EDW47477.1 CM000362 CM002911 EDX06692.1 KMY93071.1 CH479223 EDW35017.1 CH902619 EDV36743.1 AE013599 AAF58598.1 AHN56122.1 AY122236 AAM52748.1 CP012524 ALC40728.1 CM000158 EDW90858.1 GDIQ01088905 JAN05832.1 GABZ01006579 JAA46946.1 CVRI01000047 CRK97720.1 KQ978834 KYN28021.1 CH954179 EDV56403.1
KQ459602 KPI93292.1 AK402018 BAM18640.1 AGBW02014057 OWR42336.1 JN030492 AEI27594.1 PYGN01001923 PSN32127.1 GEZM01025721 JAV87371.1 KQ971343 EFA03514.2 AXCN02001464 APCN01000278 AAAB01008859 EAA08059.4 GFDL01008971 JAV26074.1 ATLV01020849 KE525308 KFB45858.1 GECL01002884 JAP03240.1 CH478095 EAT33981.1 NEVH01019374 PNF22625.1 GDKW01003201 JAI53394.1 ACPB03015003 GAHY01002059 JAA75451.1 GGFK01010479 MBW43800.1 GGFK01010510 MBW43831.1 DS232463 EDS42159.1 GGFJ01006717 MBW55858.1 ADMH02002014 ETN59988.1 UFQS01000046 UFQT01000046 SSW98524.1 SSX18910.1 AXCM01000472 GFDF01004271 JAV09813.1 GGFJ01006721 MBW55862.1 GFDF01004270 JAV09814.1 KB632364 ERL93545.1 KK852880 KDR14418.1 GAPW01002137 JAC11461.1 JXUM01080774 KQ563206 KXJ74289.1 KQ978379 KYM94488.1 JRES01000204 KNC33302.1 GL888081 EGI67898.1 KQ982294 KYQ58395.1 AJVK01016832 ADTU01016577 GEDC01004190 JAS33108.1 KQ976491 KYM83321.1 CCAG010009454 CH940662 EDW58730.1 CH916367 EDW01530.1 KRF78436.1 CH933808 EDW08425.1 GBXI01003115 JAD11177.1 CH963894 EDW77209.2 DS235044 EEB10902.1 GDHF01002670 JAI49644.1 GL434950 EFN74267.1 QOIP01000010 RLU18171.1 KK854259 PTY13848.1 CM000071 KRT02699.1 EAL25960.2 GAKP01018239 JAC40713.1 AEFK01178545 AEFK01178546 AEFK01178547 GAMC01006996 JAB99559.1 GDHC01003989 JAQ14640.1 CH480816 EDW47477.1 CM000362 CM002911 EDX06692.1 KMY93071.1 CH479223 EDW35017.1 CH902619 EDV36743.1 AE013599 AAF58598.1 AHN56122.1 AY122236 AAM52748.1 CP012524 ALC40728.1 CM000158 EDW90858.1 GDIQ01088905 JAN05832.1 GABZ01006579 JAA46946.1 CVRI01000047 CRK97720.1 KQ978834 KYN28021.1 CH954179 EDV56403.1
Proteomes
UP000283053
UP000053240
UP000218220
UP000053268
UP000007151
UP000245037
+ More
UP000007266 UP000075884 UP000075885 UP000075886 UP000075901 UP000075903 UP000076408 UP000075902 UP000075840 UP000075881 UP000075882 UP000076407 UP000007062 UP000075900 UP000030765 UP000008820 UP000235965 UP000015103 UP000002320 UP000075920 UP000000673 UP000069272 UP000075880 UP000075883 UP000192223 UP000030742 UP000027135 UP000069940 UP000249989 UP000078542 UP000037069 UP000007755 UP000075809 UP000092462 UP000095301 UP000095300 UP000005205 UP000091820 UP000092445 UP000078540 UP000092444 UP000008792 UP000001070 UP000078200 UP000009192 UP000007798 UP000009046 UP000000311 UP000279307 UP000001819 UP000007648 UP000001292 UP000000304 UP000008744 UP000192221 UP000007801 UP000000803 UP000265120 UP000092553 UP000002282 UP000221080 UP000183832 UP000192224 UP000078492 UP000008711
UP000007266 UP000075884 UP000075885 UP000075886 UP000075901 UP000075903 UP000076408 UP000075902 UP000075840 UP000075881 UP000075882 UP000076407 UP000007062 UP000075900 UP000030765 UP000008820 UP000235965 UP000015103 UP000002320 UP000075920 UP000000673 UP000069272 UP000075880 UP000075883 UP000192223 UP000030742 UP000027135 UP000069940 UP000249989 UP000078542 UP000037069 UP000007755 UP000075809 UP000092462 UP000095301 UP000095300 UP000005205 UP000091820 UP000092445 UP000078540 UP000092444 UP000008792 UP000001070 UP000078200 UP000009192 UP000007798 UP000009046 UP000000311 UP000279307 UP000001819 UP000007648 UP000001292 UP000000304 UP000008744 UP000192221 UP000007801 UP000000803 UP000265120 UP000092553 UP000002282 UP000221080 UP000183832 UP000192224 UP000078492 UP000008711
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A3S2LWI9
A0A194RCA1
A0A2A4JPJ0
A0A194PIZ5
I4DL50
A0A212ELH1
+ More
G3JWY3 A0A2P8XJE4 A0A1Y1MNY9 D6WL40 A0A182NRE1 A0A182PI63 A0A182QPL7 A0A182STB1 A0A182V8Q2 A0A182Y6L5 A0A182TQ58 A0A182HSG9 A0A182KCN9 A0A182LEH8 A0A182XAM8 Q7QCH5 A0A1Q3FEU1 A0A182RXQ3 A0A084W6L4 A0A0V0G6C3 Q16I97 A0A2J7Q205 A0A0P4VLT5 R4FLP2 A0A2M4ASR3 A0A2M4ASS3 B0X7U0 A0A2M4BSW3 A0A182W0T2 W5J9B7 A0A182FPB9 A0A182IL98 A0A336K0Q3 A0A182M9Q5 A0A1L8DTM3 A0A1W4WZ51 A0A2M4BS76 A0A1L8DTW0 U4UTS1 A0A067QYA2 A0A023ERX6 A0A182GQK3 A0A195C2S4 A0A0L0CM32 F4WD26 A0A151XDF7 A0A1B0DMJ2 A0A1I8N9M6 A0A1I8QA46 A0A158NID5 A0A1B6E5C0 A0A1A9WBA4 A0A1B0A7I5 A0A195BFG7 A0A1B0FQP0 B4MDW5 B4J3U7 A0A1A9UY58 A0A0Q9WAZ1 B4KSE3 A0A0A1XJQ2 B4MYM0 E0VBZ6 A0A0K8WF12 E1ZX17 A0A3L8DCW6 A0A2R7W342 A0A0R3NP36 Q28YK6 A0A034VGR2 G3WN22 A0A1I8QA10 W8C932 A0A146M7U5 B4HNW2 B4QC61 B4H8I2 A0A1W4UNP1 B3MG48 A1Z8R8 A0A3P8WEV8 Q8MQY3 A0A0M4E448 B4P5R1 A0A0P6CNX2 K9IXI8 A0A2D0STN9 A0A1J1IBW2 A0A1W4Z3M9 A0A151JNT4 B3NSA1
G3JWY3 A0A2P8XJE4 A0A1Y1MNY9 D6WL40 A0A182NRE1 A0A182PI63 A0A182QPL7 A0A182STB1 A0A182V8Q2 A0A182Y6L5 A0A182TQ58 A0A182HSG9 A0A182KCN9 A0A182LEH8 A0A182XAM8 Q7QCH5 A0A1Q3FEU1 A0A182RXQ3 A0A084W6L4 A0A0V0G6C3 Q16I97 A0A2J7Q205 A0A0P4VLT5 R4FLP2 A0A2M4ASR3 A0A2M4ASS3 B0X7U0 A0A2M4BSW3 A0A182W0T2 W5J9B7 A0A182FPB9 A0A182IL98 A0A336K0Q3 A0A182M9Q5 A0A1L8DTM3 A0A1W4WZ51 A0A2M4BS76 A0A1L8DTW0 U4UTS1 A0A067QYA2 A0A023ERX6 A0A182GQK3 A0A195C2S4 A0A0L0CM32 F4WD26 A0A151XDF7 A0A1B0DMJ2 A0A1I8N9M6 A0A1I8QA46 A0A158NID5 A0A1B6E5C0 A0A1A9WBA4 A0A1B0A7I5 A0A195BFG7 A0A1B0FQP0 B4MDW5 B4J3U7 A0A1A9UY58 A0A0Q9WAZ1 B4KSE3 A0A0A1XJQ2 B4MYM0 E0VBZ6 A0A0K8WF12 E1ZX17 A0A3L8DCW6 A0A2R7W342 A0A0R3NP36 Q28YK6 A0A034VGR2 G3WN22 A0A1I8QA10 W8C932 A0A146M7U5 B4HNW2 B4QC61 B4H8I2 A0A1W4UNP1 B3MG48 A1Z8R8 A0A3P8WEV8 Q8MQY3 A0A0M4E448 B4P5R1 A0A0P6CNX2 K9IXI8 A0A2D0STN9 A0A1J1IBW2 A0A1W4Z3M9 A0A151JNT4 B3NSA1
Ontologies
GO
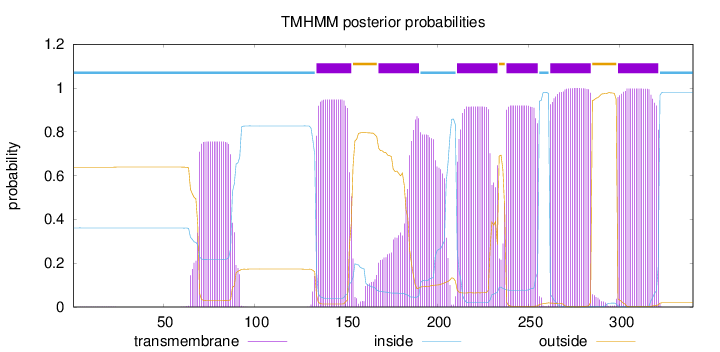
Topology
Length:
340
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
137.90915
Exp number, first 60 AAs:
0.0142
Total prob of N-in:
0.36127
inside
1 - 133
TMhelix
134 - 153
outside
154 - 167
TMhelix
168 - 190
inside
191 - 210
TMhelix
211 - 233
outside
234 - 237
TMhelix
238 - 255
inside
256 - 261
TMhelix
262 - 284
outside
285 - 298
TMhelix
299 - 321
inside
322 - 340
Population Genetic Test Statistics
Pi
269.576118
Theta
201.09313
Tajima's D
1.205722
CLR
1.353305
CSRT
0.713814309284536
Interpretation
Uncertain
