Gene
KWMTBOMO08926 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007720
Annotation
PREDICTED:_serine_protease_inhibitor_2_isoform_X1_[Bombyx_mori]
Full name
Serine protease inhibitor 3/4
+ More
Antitrypsin
Antichymotrypsin-2
Alaserpin
Antitrypsin
Antichymotrypsin-2
Alaserpin
Alternative Name
Antichymotrypsin II
Serpin-1
Serpin-1
Location in the cell
Cytoplasmic Reliability : 1.041 Extracellular Reliability : 1.107
Sequence
CDS
ATGATATTTGCCAGAGTTTTGTTAATACTATTTACACTATTGGAAATAAAGAACCCTGTTCTCAGTATGGATTCAAAGGCTTTATCCTCTGCAATCACCAAGTTCTCTGCAAAGTTTTGCAATGAACTGGATAAAAAGAAGAATGTTGTATCATCGCCATTGTCAGCAGAATACTTGCTGGCGTTGATAACTTTGGGAACTACTGATCCTGCACACGAAGAGCTATTGACAAGCCTAGGTATCCCGGATGATGACACGATTCGCTCATCATTCTCGGCTGTGTCGTCAAAATTGAAATCAATAAAAGGTGTTACGTTTAATGTAGCCAACAAAATATACATCAAGGAGGGTGACTATGAGCTCGACCCAAAACTTAAGAAGGACGCTGTCGAGGTATTCGATGCAGATTTTGAAAAAGTTAACTTCGATAACGGAGCCGCAGCCGCCGGCCTAATAAACAAATGGGTTGAAAATAAAACAAATGAACGCATTAAAGATCTCTTGAGTGAAGACAGCCTCGATTCGTTTACGCGTTTGGTTTTGGTCAACGCTCTCTACTTCAAGGGTACATGGCAAAACCAGTTCGACTCTATAAGCACCATGGAGCGACCGTTCTATGTTGACACCGAAACGACAGTTAATATTCCTATGATGTACCAAGAAAATAATTTCAAGTATGGTGAGAGCCACGACCTCAATGCGCAGTTACTCGAAATGGCGTATGAAGGTAACGATGCCAGCATGGTTATCGTGCTGCCGAACGAAATTAATGGCCTGGACGGGATACTGCAGAAGCTGGCCGATGGCTACGACCTAACGGCGGAACTGGACAAAATGTTCTCGACCAAAGTGCGAGTGACGGTGCCGAAATTCAAGATCGAAACTGAGATTGATCTACTTCAAGTTTTGCCCAAGTTGGGTATTCAGGCGATCTTCAACCGCCAGAATTCCGGTCTGACCAAGATCTTGGATAACGACGAGCCGCTGTACGTATCCAAGGCTGTGCAAAAAGCCTTCATTGAAGTCAACGAGGAAGGCGCCGAAGCAGCCGCCGCCACAGCAATGGGCATTGCAAGTTACGCTGCATTCCTTGATGAGCCACCAGTACCTTCCTTCGATGCTGATCGGCCATTTGTAGCCGCAATCTTTGCCCACGGAATCTCTCACTTCTATGCCATATACCGTGGGGAGAATGATGAATAA
Protein
MIFARVLLILFTLLEIKNPVLSMDSKALSSAITKFSAKFCNELDKKKNVVSSPLSAEYLLALITLGTTDPAHEELLTSLGIPDDDTIRSSFSAVSSKLKSIKGVTFNVANKIYIKEGDYELDPKLKKDAVEVFDADFEKVNFDNGAAAAGLINKWVENKTNERIKDLLSEDSLDSFTRLVLVNALYFKGTWQNQFDSISTMERPFYVDTETTVNIPMMYQENNFKYGESHDLNAQLLEMAYEGNDASMVIVLPNEINGLDGILQKLADGYDLTAELDKMFSTKVRVTVPKFKIETEIDLLQVLPKLGIQAIFNRQNSGLTKILDNDEPLYVSKAVQKAFIEVNEEGAEAAAATAMGIASYAAFLDEPPVPSFDADRPFVAAIFAHGISHFYAIYRGENDE
Summary
Miscellaneous
The N-terminal section (1-336) is identical to the N-terminal section of the silk moth antitrypsin I (17-352).
Similarity
Belongs to the serpin family.
Keywords
Alternative splicing
Direct protein sequencing
Protease inhibitor
Serine protease inhibitor
Signal
Complete proteome
Reference proteome
Secreted
3D-structure
Glycoprotein
Feature
chain Serine protease inhibitor 3/4
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A0M4JBR8
Q9NGS0
E2GHX4
H9JDX3
A0A2A4JPB5
A0A2A4JQC6
+ More
B2X122 A0A2A4JR25 A0A2H1VAC5 A0A2A4JPH1 A0A194PIN8 Q5MGH1 J7HI16 A0A1L7B973 Q5MGH2 A0A2A4JQ63 C0J8H0 A0A1E1WB01 A0A3S2NC81 A0A1E1W229 A0A212EXP2 A0A1L7B974 A0A212ELH5 A0A1E1W071 A0A437BU62 G9F9J6 O02377 A3KEZ8 A0A2H1VE36 C7ASM4 Q5MGH0 C7ASM6 A3KEZ7 Q60FS0 B4XHA1 B4XH99 B4XHA0 F5B4G8 A0A0M4K7J2 A0A1W4XA17 A0A1W4X0N9 C7ASM3 C7ASM7 A0A1W4XB66 C7ASM2 P22922 C7ASM9 P80034 A0A1W4XA13 A0A1W4X0K9 A0A1W4XB61 A0A3S2M7J7 A0A3G1T1E7 A0A1W4X0J8 A0A1W4XBF6 A0A1W4XA23 A0A1L7B975 K4P392 A0A1W4XBF1 A0A1W4XBE6 A0A194R1A3 B4XH98 H9JKA2 Q25496 A0A212FD15 Q5MGH0-2 Q8IS84 Q25501 Q25495 A0A1Y1KQA5 A0A385XRT7 I4DJC8 Q25500 A0A194PZ13 Q25497 D9HQA1 B0FZ62 G9D4T8 C5I795 A0A385XRR7 A0A1Y1KQA4 B0FZ64 A0A1Y1KTQ8 Q25492 B0FZ60 A0A1Y1KVX7 Q8IS86 C5I794 B0FZ63 A0A1Y1KX43 B0FZ65 B0FZ67 P14754 Q25526
B2X122 A0A2A4JR25 A0A2H1VAC5 A0A2A4JPH1 A0A194PIN8 Q5MGH1 J7HI16 A0A1L7B973 Q5MGH2 A0A2A4JQ63 C0J8H0 A0A1E1WB01 A0A3S2NC81 A0A1E1W229 A0A212EXP2 A0A1L7B974 A0A212ELH5 A0A1E1W071 A0A437BU62 G9F9J6 O02377 A3KEZ8 A0A2H1VE36 C7ASM4 Q5MGH0 C7ASM6 A3KEZ7 Q60FS0 B4XHA1 B4XH99 B4XHA0 F5B4G8 A0A0M4K7J2 A0A1W4XA17 A0A1W4X0N9 C7ASM3 C7ASM7 A0A1W4XB66 C7ASM2 P22922 C7ASM9 P80034 A0A1W4XA13 A0A1W4X0K9 A0A1W4XB61 A0A3S2M7J7 A0A3G1T1E7 A0A1W4X0J8 A0A1W4XBF6 A0A1W4XA23 A0A1L7B975 K4P392 A0A1W4XBF1 A0A1W4XBE6 A0A194R1A3 B4XH98 H9JKA2 Q25496 A0A212FD15 Q5MGH0-2 Q8IS84 Q25501 Q25495 A0A1Y1KQA5 A0A385XRT7 I4DJC8 Q25500 A0A194PZ13 Q25497 D9HQA1 B0FZ62 G9D4T8 C5I795 A0A385XRR7 A0A1Y1KQA4 B0FZ64 A0A1Y1KTQ8 Q25492 B0FZ60 A0A1Y1KVX7 Q8IS86 C5I794 B0FZ63 A0A1Y1KX43 B0FZ65 B0FZ67 P14754 Q25526
Pubmed
EMBL
KR003725
ALD62503.1
AF242200
AAF61252.1
HM357255
ADN52338.1
+ More
BABH01002639 BABH01002640 NWSH01000840 PCG73905.1 PCG73908.1 EF584499 ABU62829.1 PCG73903.1 ODYU01001314 SOQ37342.1 PCG73901.1 KQ459602 KPI93286.1 AY829815 AAV91429.1 JN400442 AFQ01141.1 KY094503 APT69915.1 AY829814 AAV91428.1 PCG73906.1 EU935622 ACG61184.1 GDQN01006900 JAT84154.1 RSAL01000236 RVE43711.1 GDQN01010075 GDQN01007015 JAT80979.1 JAT84039.1 AGBW02011729 OWR46227.1 KY385636 APT69917.1 AGBW02014057 OWR42342.1 GDQN01010691 JAT80363.1 RSAL01000008 RVE53905.1 JN033741 AEW46894.1 U79184 AAB58491.1 AB295499 BAF48335.1 ODYU01002062 SOQ39103.1 FJ613795 ACT36274.1 AY829816 AY829817 AAV91430.1 FJ613797 ACT36278.1 AB295498 BAF48334.1 AB189029 BAD52261.1 EU040210 ABW17158.1 EU040208 ABW17156.1 EU040209 ABW17157.1 HQ615869 ADT63775.1 KR003729 ALD62507.1 FJ613794 FJ613796 ACT36273.1 ACT36275.1 ACT36277.1 ACT36279.1 FJ613793 ACT36272.1 D00738 ACT36276.1 RSAL01000018 RVE52837.1 MG992370 AXY94808.1 KY385635 APT69916.1 JX524148 AFV46312.1 KQ460883 KPJ11294.1 EU040207 ABW17155.1 BABH01041150 U58361 AAC47336.1 AGBW02009121 OWR51607.1 AY148485 AAN71634.1 AAC47339.1 AAC47341.1 GEZM01076851 GEZM01076849 JAV63593.1 MG753798 AYC12555.1 AK401396 BAM18018.1 AAC47335.1 KQ459586 KPI97989.1 AAC47333.1 HM023856 ADJ58589.2 EU368832 ABY68561.1 JF777341 AEU11686.1 FJ763764 ACR56865.1 MG753799 AYC12556.1 GEZM01076850 JAV63592.1 ABY68559.1 GEZM01076852 JAV63590.1 AAC47338.1 ABY68563.1 GEZM01073936 JAV64728.1 AY148483 AAN71632.1 FJ763763 ACR56864.1 ABY68560.1 GEZM01073934 JAV64730.1 ABY68558.1 ABY68556.1 M23438 L20792 L20793 AAA29334.1
BABH01002639 BABH01002640 NWSH01000840 PCG73905.1 PCG73908.1 EF584499 ABU62829.1 PCG73903.1 ODYU01001314 SOQ37342.1 PCG73901.1 KQ459602 KPI93286.1 AY829815 AAV91429.1 JN400442 AFQ01141.1 KY094503 APT69915.1 AY829814 AAV91428.1 PCG73906.1 EU935622 ACG61184.1 GDQN01006900 JAT84154.1 RSAL01000236 RVE43711.1 GDQN01010075 GDQN01007015 JAT80979.1 JAT84039.1 AGBW02011729 OWR46227.1 KY385636 APT69917.1 AGBW02014057 OWR42342.1 GDQN01010691 JAT80363.1 RSAL01000008 RVE53905.1 JN033741 AEW46894.1 U79184 AAB58491.1 AB295499 BAF48335.1 ODYU01002062 SOQ39103.1 FJ613795 ACT36274.1 AY829816 AY829817 AAV91430.1 FJ613797 ACT36278.1 AB295498 BAF48334.1 AB189029 BAD52261.1 EU040210 ABW17158.1 EU040208 ABW17156.1 EU040209 ABW17157.1 HQ615869 ADT63775.1 KR003729 ALD62507.1 FJ613794 FJ613796 ACT36273.1 ACT36275.1 ACT36277.1 ACT36279.1 FJ613793 ACT36272.1 D00738 ACT36276.1 RSAL01000018 RVE52837.1 MG992370 AXY94808.1 KY385635 APT69916.1 JX524148 AFV46312.1 KQ460883 KPJ11294.1 EU040207 ABW17155.1 BABH01041150 U58361 AAC47336.1 AGBW02009121 OWR51607.1 AY148485 AAN71634.1 AAC47339.1 AAC47341.1 GEZM01076851 GEZM01076849 JAV63593.1 MG753798 AYC12555.1 AK401396 BAM18018.1 AAC47335.1 KQ459586 KPI97989.1 AAC47333.1 HM023856 ADJ58589.2 EU368832 ABY68561.1 JF777341 AEU11686.1 FJ763764 ACR56865.1 MG753799 AYC12556.1 GEZM01076850 JAV63592.1 ABY68559.1 GEZM01076852 JAV63590.1 AAC47338.1 ABY68563.1 GEZM01073936 JAV64728.1 AY148483 AAN71632.1 FJ763763 ACR56864.1 ABY68560.1 GEZM01073934 JAV64730.1 ABY68558.1 ABY68556.1 M23438 L20792 L20793 AAA29334.1
Proteomes
Pfam
PF00079 Serpin
Interpro
SUPFAM
SSF56574
SSF56574
Gene 3D
ProteinModelPortal
A0A0M4JBR8
Q9NGS0
E2GHX4
H9JDX3
A0A2A4JPB5
A0A2A4JQC6
+ More
B2X122 A0A2A4JR25 A0A2H1VAC5 A0A2A4JPH1 A0A194PIN8 Q5MGH1 J7HI16 A0A1L7B973 Q5MGH2 A0A2A4JQ63 C0J8H0 A0A1E1WB01 A0A3S2NC81 A0A1E1W229 A0A212EXP2 A0A1L7B974 A0A212ELH5 A0A1E1W071 A0A437BU62 G9F9J6 O02377 A3KEZ8 A0A2H1VE36 C7ASM4 Q5MGH0 C7ASM6 A3KEZ7 Q60FS0 B4XHA1 B4XH99 B4XHA0 F5B4G8 A0A0M4K7J2 A0A1W4XA17 A0A1W4X0N9 C7ASM3 C7ASM7 A0A1W4XB66 C7ASM2 P22922 C7ASM9 P80034 A0A1W4XA13 A0A1W4X0K9 A0A1W4XB61 A0A3S2M7J7 A0A3G1T1E7 A0A1W4X0J8 A0A1W4XBF6 A0A1W4XA23 A0A1L7B975 K4P392 A0A1W4XBF1 A0A1W4XBE6 A0A194R1A3 B4XH98 H9JKA2 Q25496 A0A212FD15 Q5MGH0-2 Q8IS84 Q25501 Q25495 A0A1Y1KQA5 A0A385XRT7 I4DJC8 Q25500 A0A194PZ13 Q25497 D9HQA1 B0FZ62 G9D4T8 C5I795 A0A385XRR7 A0A1Y1KQA4 B0FZ64 A0A1Y1KTQ8 Q25492 B0FZ60 A0A1Y1KVX7 Q8IS86 C5I794 B0FZ63 A0A1Y1KX43 B0FZ65 B0FZ67 P14754 Q25526
B2X122 A0A2A4JR25 A0A2H1VAC5 A0A2A4JPH1 A0A194PIN8 Q5MGH1 J7HI16 A0A1L7B973 Q5MGH2 A0A2A4JQ63 C0J8H0 A0A1E1WB01 A0A3S2NC81 A0A1E1W229 A0A212EXP2 A0A1L7B974 A0A212ELH5 A0A1E1W071 A0A437BU62 G9F9J6 O02377 A3KEZ8 A0A2H1VE36 C7ASM4 Q5MGH0 C7ASM6 A3KEZ7 Q60FS0 B4XHA1 B4XH99 B4XHA0 F5B4G8 A0A0M4K7J2 A0A1W4XA17 A0A1W4X0N9 C7ASM3 C7ASM7 A0A1W4XB66 C7ASM2 P22922 C7ASM9 P80034 A0A1W4XA13 A0A1W4X0K9 A0A1W4XB61 A0A3S2M7J7 A0A3G1T1E7 A0A1W4X0J8 A0A1W4XBF6 A0A1W4XA23 A0A1L7B975 K4P392 A0A1W4XBF1 A0A1W4XBE6 A0A194R1A3 B4XH98 H9JKA2 Q25496 A0A212FD15 Q5MGH0-2 Q8IS84 Q25501 Q25495 A0A1Y1KQA5 A0A385XRT7 I4DJC8 Q25500 A0A194PZ13 Q25497 D9HQA1 B0FZ62 G9D4T8 C5I795 A0A385XRR7 A0A1Y1KQA4 B0FZ64 A0A1Y1KTQ8 Q25492 B0FZ60 A0A1Y1KVX7 Q8IS86 C5I794 B0FZ63 A0A1Y1KX43 B0FZ65 B0FZ67 P14754 Q25526
PDB
1SEK
E-value=2.19607e-58,
Score=571
Ontologies
PANTHER
Topology
Subcellular location
Secreted
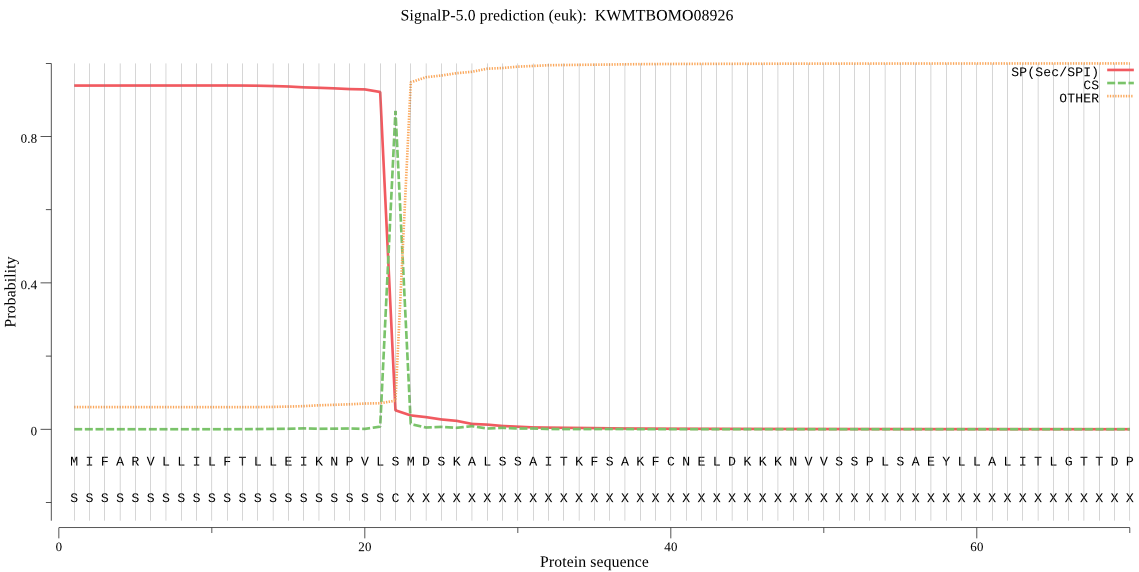
SignalP
Position: 1 - 22,
Likelihood: 0.939161
Length:
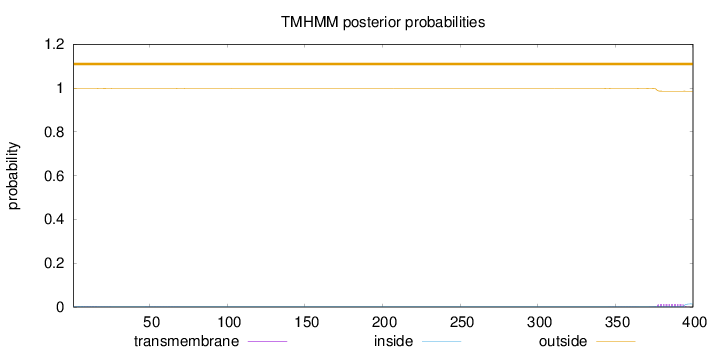
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28244
Exp number, first 60 AAs:
0.03039
Total prob of N-in:
0.00294
outside
1 - 400
Population Genetic Test Statistics
Pi
193.738088
Theta
167.608042
Tajima's D
-1.445339
CLR
10.476372
CSRT
0.0689965501724914
Interpretation
Uncertain
