Gene
KWMTBOMO08924
Pre Gene Modal
BGIBMGA007812
Annotation
PREDICTED:_regulating_synaptic_membrane_exocytosis_protein_2_isoform_X5_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.717
Sequence
CDS
ATGCCGGTATCATCTCGAACAATTCCTCAGAGCACTCCGGTACAAAATACAGAGGGAACCGAAGTCCCTCCGCAGACCCAGAGGTTTCAAACGATCCGTAAGAATGGGATTATCGACAATCCGAACGGCCCTCCTCTGTATGGAGTCAAATGGAAGAAGCTCGTATTTGGAAGCCCCGGCCCAGAGATGGGAGCAGTACTCCACGCGAGGCCGGACTTGTGCTTTATAAAGCAAAAGTCTTTGTTCAGGCGTGAAGTACCGCTTCGCTCTGTTGAGGACTCCCAGCATTTTGGACGCCAACTTGGCTTTGCCTTCCAAATGACTCAGAAACTGGACATCGCTCGAAATGTCGACCCCAAGTATCCCGATACTCTCGGAAGGTTGCAGGGATACTTCTTGGAATTGCGGCGCCATGACAAAGGGGTCCTTCTTCGCAGATGGGAGCGACATTATGCGGGTTTATCTGATTCGGAGCTTGCTGCCCGCAGTGGCGGTAGTTGGGCTCCACGGAGAAGGCTATCTCCTGATGCAACAGCAGCAGATTCCGACCTAGAATCGGTCGCTTCTGTAACATCAAGCGCCTTCAGCACACAATCCGAACGACCTCGACCGACACGTATGCTTAGCAACGATTATGGTGGAAGGGACAACGGAAGGAACAATAATCAGCCACAGCCGTTGGAGAGACGCGAGTCTCGCCGAGGCCAATTCACTCGGAGTTTGAGCAACGCAGATGTGCCGCCTGATGAAAAAACAGATGGCAGTCTAAGTGACACTGCGTTGGGTGGTACCGGCGAGCTGGAGCCAAGCACTGATGATGTCCTCTTAAAAGCTGAGCCTCGGGATTATTTCGGGCCGGGCATGGGCAAGAAAAGCAACTCTACCTCACAATTGTCAGCCACAGGACGAAAACGAAGGTTAGGTTTTGGTAAACGTGGCAAAAATTCGTTTACGGTGCAACGCAGTGAGGAAGTGGTACCCGGCGAAATGAGAGGAGGCACCGGCGTGTCGCGGGCCTCCTCCGCATCCAGTGAGAACGACGAAGACAGGTGGTCCCCGGGCATGCGAGGTTCTGGTTCTTCAGAAGGCGGAGGGCTATCGGACTTCATTGACGGCCTGGGTCCCGGACAGCTGGTCGGGAGACAATTACTAGGGGCGACTACGCTTGGAGATGTCCAACTTTCAATGTGCTACCAAAAAGGATTCTTAGAGGTGGAAGTCATACGTGCTCGCGGCTTACAAGCCAGGCAAGGAAGCCGAACCCTTCCAGCGCCATACGTCAAGGTCTATCTCGTGAGCGGAAAGCGATGTATAGCCAAAGCAAAGACAACAACCGCACGACGTACGCTCGACCCTCTCTACCAGCAAACGCTAACGTTTCGAGAAAACTTCAAAGGATGTGTACTGCAGGTTACTGTTTGGGGTGACTACGGTCGCATCGAGGGGAAAAAAGTATTTATGGGCGTGGCCCAAATAATGCTGGACGACATAAACCTCTCTAATATCGTTATTGGATGGTACAAACTATTTGGAACTACTTCTTTGTAA
Protein
MPVSSRTIPQSTPVQNTEGTEVPPQTQRFQTIRKNGIIDNPNGPPLYGVKWKKLVFGSPGPEMGAVLHARPDLCFIKQKSLFRREVPLRSVEDSQHFGRQLGFAFQMTQKLDIARNVDPKYPDTLGRLQGYFLELRRHDKGVLLRRWERHYAGLSDSELAARSGGSWAPRRRLSPDATAADSDLESVASVTSSAFSTQSERPRPTRMLSNDYGGRDNGRNNNQPQPLERRESRRGQFTRSLSNADVPPDEKTDGSLSDTALGGTGELEPSTDDVLLKAEPRDYFGPGMGKKSNSTSQLSATGRKRRLGFGKRGKNSFTVQRSEEVVPGEMRGGTGVSRASSASSENDEDRWSPGMRGSGSSEGGGLSDFIDGLGPGQLVGRQLLGATTLGDVQLSMCYQKGFLEVEVIRARGLQARQGSRTLPAPYVKVYLVSGKRCIAKAKTTTARRTLDPLYQQTLTFRENFKGCVLQVTVWGDYGRIEGKKVFMGVAQIMLDDINLSNIVIGWYKLFGTTSL
Summary
Uniprot
A0A437BIY2
A0A2J7QL53
A0A2J7QL47
A0A1Y1N8Q6
A0A1Y1N9T5
A0A1J1J266
+ More
A0A1B6CKH8 A0A1B6CLW7 A0A1W4WTF2 A0A1Y1N6A6 D6WLW5 A0A1Y1N537 A0A2J7QL51 A0A0A9Y1U9 E0VWR1 T1H9W1 A0A336LKE1 A0A336LP26 A0A0A9VVP2 A0A146KXX4 A0A0A9Y4V6 A0A0A9VVP8 A0A0A9W1H3 A0A0A9VYQ8 V9IBN9 A0A0Q9WS98 A0A0Q9WR78 A0A0Q9WQ16 U4U462 A0A0A9XZA4 A0A146M6M7 A0A0A9W1G8 A0A0A9W622 A0A0A9W627 A0A0A9VXD4 A0A0A9VXC8 A0A0Q9WXZ7 A0A0Q9WX46 A0A0Q9X9N8 A0A0Q9X7V4 F4W8W8 A0A0B4K677 A0A0R1E3F8 A0A0B4K7E1 Q9VEE5 A0A1W4UJA5 A0A0R3NQU1 A0A0R3NPH2
A0A1B6CKH8 A0A1B6CLW7 A0A1W4WTF2 A0A1Y1N6A6 D6WLW5 A0A1Y1N537 A0A2J7QL51 A0A0A9Y1U9 E0VWR1 T1H9W1 A0A336LKE1 A0A336LP26 A0A0A9VVP2 A0A146KXX4 A0A0A9Y4V6 A0A0A9VVP8 A0A0A9W1H3 A0A0A9VYQ8 V9IBN9 A0A0Q9WS98 A0A0Q9WR78 A0A0Q9WQ16 U4U462 A0A0A9XZA4 A0A146M6M7 A0A0A9W1G8 A0A0A9W622 A0A0A9W627 A0A0A9VXD4 A0A0A9VXC8 A0A0Q9WXZ7 A0A0Q9WX46 A0A0Q9X9N8 A0A0Q9X7V4 F4W8W8 A0A0B4K677 A0A0R1E3F8 A0A0B4K7E1 Q9VEE5 A0A1W4UJA5 A0A0R3NQU1 A0A0R3NPH2
Pubmed
EMBL
RSAL01000049
RVE50400.1
NEVH01013250
PNF29315.1
PNF29314.1
GEZM01012342
+ More
JAV92985.1 GEZM01012341 JAV92986.1 CVRI01000066 CRL06042.1 GEDC01023410 JAS13888.1 GEDC01022876 JAS14422.1 GEZM01012339 JAV92988.1 KQ971343 EFA04175.2 GEZM01012344 JAV92983.1 PNF29313.1 GBHO01017480 JAG26124.1 DS235823 EEB17817.1 ACPB03014673 ACPB03014674 ACPB03014675 UFQT01000038 SSX18426.1 SSX18423.1 GBHO01043297 JAG00307.1 GDHC01017336 JAQ01293.1 GBHO01017481 JAG26123.1 GBHO01043292 JAG00312.1 GBHO01043291 JAG00313.1 GBHO01043293 JAG00311.1 JR039064 AEY58515.1 CH940650 KRF83109.1 KRF83115.1 KRF83119.1 KB632094 ERL88699.1 GBHO01017482 JAG26122.1 GDHC01004202 JAQ14427.1 GBHO01043296 JAG00308.1 GBHO01043294 JAG00310.1 GBHO01043289 JAG00315.1 GBHO01043290 JAG00314.1 GBHO01043295 JAG00309.1 CH933806 KRG00613.1 KRG00614.1 KRG00611.1 KRG00631.1 GL887973 EGI69360.1 AE014297 AFH06482.2 CM000160 KRK02755.1 AFH06479.2 AFH06484.2 AAF55479.5 CM000070 KRT01296.1 KRT01295.1
JAV92985.1 GEZM01012341 JAV92986.1 CVRI01000066 CRL06042.1 GEDC01023410 JAS13888.1 GEDC01022876 JAS14422.1 GEZM01012339 JAV92988.1 KQ971343 EFA04175.2 GEZM01012344 JAV92983.1 PNF29313.1 GBHO01017480 JAG26124.1 DS235823 EEB17817.1 ACPB03014673 ACPB03014674 ACPB03014675 UFQT01000038 SSX18426.1 SSX18423.1 GBHO01043297 JAG00307.1 GDHC01017336 JAQ01293.1 GBHO01017481 JAG26123.1 GBHO01043292 JAG00312.1 GBHO01043291 JAG00313.1 GBHO01043293 JAG00311.1 JR039064 AEY58515.1 CH940650 KRF83109.1 KRF83115.1 KRF83119.1 KB632094 ERL88699.1 GBHO01017482 JAG26122.1 GDHC01004202 JAQ14427.1 GBHO01043296 JAG00308.1 GBHO01043294 JAG00310.1 GBHO01043289 JAG00315.1 GBHO01043290 JAG00314.1 GBHO01043295 JAG00309.1 CH933806 KRG00613.1 KRG00614.1 KRG00611.1 KRG00631.1 GL887973 EGI69360.1 AE014297 AFH06482.2 CM000160 KRK02755.1 AFH06479.2 AFH06484.2 AAF55479.5 CM000070 KRT01296.1 KRT01295.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A437BIY2
A0A2J7QL53
A0A2J7QL47
A0A1Y1N8Q6
A0A1Y1N9T5
A0A1J1J266
+ More
A0A1B6CKH8 A0A1B6CLW7 A0A1W4WTF2 A0A1Y1N6A6 D6WLW5 A0A1Y1N537 A0A2J7QL51 A0A0A9Y1U9 E0VWR1 T1H9W1 A0A336LKE1 A0A336LP26 A0A0A9VVP2 A0A146KXX4 A0A0A9Y4V6 A0A0A9VVP8 A0A0A9W1H3 A0A0A9VYQ8 V9IBN9 A0A0Q9WS98 A0A0Q9WR78 A0A0Q9WQ16 U4U462 A0A0A9XZA4 A0A146M6M7 A0A0A9W1G8 A0A0A9W622 A0A0A9W627 A0A0A9VXD4 A0A0A9VXC8 A0A0Q9WXZ7 A0A0Q9WX46 A0A0Q9X9N8 A0A0Q9X7V4 F4W8W8 A0A0B4K677 A0A0R1E3F8 A0A0B4K7E1 Q9VEE5 A0A1W4UJA5 A0A0R3NQU1 A0A0R3NPH2
A0A1B6CKH8 A0A1B6CLW7 A0A1W4WTF2 A0A1Y1N6A6 D6WLW5 A0A1Y1N537 A0A2J7QL51 A0A0A9Y1U9 E0VWR1 T1H9W1 A0A336LKE1 A0A336LP26 A0A0A9VVP2 A0A146KXX4 A0A0A9Y4V6 A0A0A9VVP8 A0A0A9W1H3 A0A0A9VYQ8 V9IBN9 A0A0Q9WS98 A0A0Q9WR78 A0A0Q9WQ16 U4U462 A0A0A9XZA4 A0A146M6M7 A0A0A9W1G8 A0A0A9W622 A0A0A9W627 A0A0A9VXD4 A0A0A9VXC8 A0A0Q9WXZ7 A0A0Q9WX46 A0A0Q9X9N8 A0A0Q9X7V4 F4W8W8 A0A0B4K677 A0A0R1E3F8 A0A0B4K7E1 Q9VEE5 A0A1W4UJA5 A0A0R3NQU1 A0A0R3NPH2
PDB
2Q3X
E-value=4.82466e-36,
Score=380
Ontologies
GO
GO:0006887
GO:0017137
GO:0016020
GO:0046872
GO:0005623
GO:0006886
GO:0016021
GO:0042391
GO:0098831
GO:0048788
GO:0050806
GO:0044325
GO:0045202
GO:2000300
GO:0042734
GO:0048167
GO:0048791
GO:0008021
GO:0048786
GO:0007274
GO:0016192
GO:0070073
GO:0016079
GO:0007269
GO:0017156
GO:0005515
GO:0007264
GO:0008270
GO:0016813
GO:0016817
GO:0043565
GO:0016876
GO:0043039
PANTHER
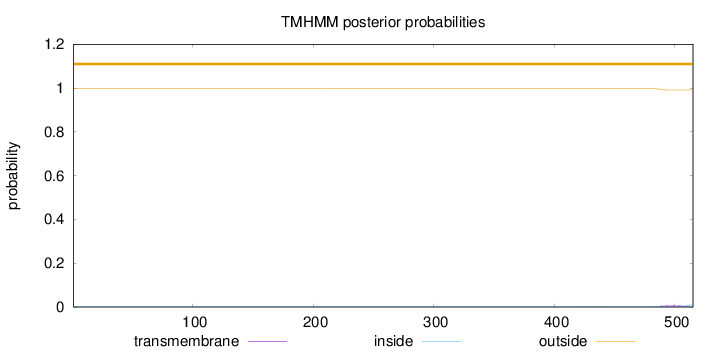
Topology
Length:
515
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16446
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.00186
outside
1 - 515
Population Genetic Test Statistics
Pi
254.356122
Theta
212.367965
Tajima's D
0.584529
CLR
0.048505
CSRT
0.542822858857057
Interpretation
Uncertain
