Gene
KWMTBOMO08923 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007719
Annotation
PREDICTED:_host_cell_factor-like_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.674
Sequence
CDS
ATGGAATCGGACCCTGCTACTCACAGCGATTCTCACGATGGCGCGGAAGTGTCGCCTCAAGCCGATAACCAGGATAACCAGGTTGCCGATACTATCGATGATTTGCCTGACAATAATTCAGCAAGTGACGATGTGACCACTGAGCCTGAGGAACTTGCTGCGAATGGTGGTCCAACTAGCAGCAGCGATGTTTTGGACGTAGAGCCTTCTATTGCACCTCCTGCTCAGGAAGAGCATCCTGTAGTTGAACAAGGTAATGCAGATGCTGAAGATGAAATGGACATAGATGAAACTGCTTCAGGTGCTATGAACGAGGAATCAGCGTATATCGGAGATAATTGTCTTTCTACACCTGCAGATACTGAAGAGAATTCGCCCCAAAATGATCAAGACAATCAACTAGATTCTATGATAAAGGAGCCAGAGTCAGAAGAAGCAGAGGGTCTAGGTGATGCTGAAGAGTCTCCTGATCAGTCTATCAGTTCTGCCATTCTACCAACAGATAGTGGTGTAGGTGAAGTTGAAGGAGCACCAAGCGTTCAAGATGATGAATCAAGTACAATGGATGAAGATATAGCTGAAGGAGGGGTTGCCAGCAGTGATGATGTCAACGACATTAGTAGTGCCGCACAGGAGATACTCAGCACTGGAATAAGCTCGAGCACTGCAGAACCTGGATCAGACATTTCAAGCAGCGGCCAAACAGAGCCGTCATTAATATCATCCACAGCCAATGGTCCAGGCATAGTACATTCGTTTTCGCTCACTCCTCAACAGCAGACCACAAACGAGTTCGGCGACGCTGATACCTCGGAGATGGAAGGCGCCGCCGACACGGCACCCGCGGGCAGTGAGTCGTCCATGCCGCTTCTCACCATGACGGCCAACGGATCCGCCGCACTCATCTCGCCGACCTCGACGCCGACGTCTACTCCGATCCAAGGCGAGGAGGGCGGGGCGACGTCGGTGTCGTCATCGGTTGGTGCGGCATGTGCGGCGGATGCGGCAAGTGATGTCGGCGCGGCGTCCGCTGCGAGCGGCGAGGCGGAAGGAGCGGTGAGCAGCACCCTCCCCTCCACCATCGCTGCGCACGCTCTTGCCACACTCGCCAGTGCTGCTCTTCACCATCACGAACAGGCCGAACCAGAAGAGCCCAAGAAACACGACGAGGACGCTTGGTACACGGTTGGCATCGTCAAAGGAACTACGTTTACGGTTCAAAATTATATATCGGATCCAAACATAGACTTGTCTAATTTATCTTTAGACAGTCTTCCCGATTTGACCGGGCTGCCGACGACGCCCTTAGAGCATGGCACAGCCTATAAGTTCAGAATTGCTGCTGTGAATTCTTGCGGCCGAGGAGAGTTCAGTGAAGAATCTGCATTCAAAACCTGTCTGCCGGGATTCCCCGGTGCGCCGTCAGCTATCAAAATATCAAAATCAGTAGAGGGCGCACATTTGTCCTGGGAACCACCGCAAGTTGCCGCCGAAGGAATTTTCGAATATTCGGTCTACCTGGCGGTGCGCTCCAACACTCAGCCTAAGGAAGCGGCGAAGTCGCAGCTGGCGTTCGTGCGCGTGTACTGTGGCAAGGCGAACACGTGCGTGGTGCCGCAGTCGTCGCTGAGTGCGGCGCACGTGGACTCCTCCACCAAGCCCGCCATCATCTTCCGCATCGCCGCGCGCAACGAGAAGGGCTACGGCCCCGCCACGCAAGTCAGGTGGCTGCAAGATATTAAATCCACTGGTGTGAAAAGAGCAGGAGAGGGAAGGTTACCGGGAGCGTCCCCATCAAAGCAACCTAAACAAATAGTACATTAA
Protein
MESDPATHSDSHDGAEVSPQADNQDNQVADTIDDLPDNNSASDDVTTEPEELAANGGPTSSSDVLDVEPSIAPPAQEEHPVVEQGNADAEDEMDIDETASGAMNEESAYIGDNCLSTPADTEENSPQNDQDNQLDSMIKEPESEEAEGLGDAEESPDQSISSAILPTDSGVGEVEGAPSVQDDESSTMDEDIAEGGVASSDDVNDISSAAQEILSTGISSSTAEPGSDISSSGQTEPSLISSTANGPGIVHSFSLTPQQQTTNEFGDADTSEMEGAADTAPAGSESSMPLLTMTANGSAALISPTSTPTSTPIQGEEGGATSVSSSVGAACAADAASDVGAASAASGEAEGAVSSTLPSTIAAHALATLASAALHHHEQAEPEEPKKHDEDAWYTVGIVKGTTFTVQNYISDPNIDLSNLSLDSLPDLTGLPTTPLEHGTAYKFRIAAVNSCGRGEFSEESAFKTCLPGFPGAPSAIKISKSVEGAHLSWEPPQVAAEGIFEYSVYLAVRSNTQPKEAAKSQLAFVRVYCGKANTCVVPQSSLSAAHVDSSTKPAIIFRIAARNEKGYGPATQVRWLQDIKSTGVKRAGEGRLPGASPSKQPKQIVH
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
SUPFAM
SSF49265
SSF49265
Gene 3D
CDD
ProteinModelPortal
PDB
4GO6
E-value=1.39245e-51,
Score=515
Ontologies
GO
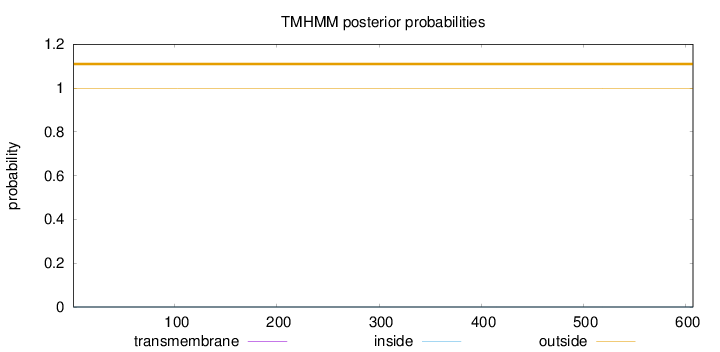
Topology
Length:
607
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00035
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00053
outside
1 - 607
Population Genetic Test Statistics
Pi
208.154762
Theta
166.864556
Tajima's D
0.692713
CLR
0.147233
CSRT
0.566671666416679
Interpretation
Uncertain
