Gene
KWMTBOMO08922
Pre Gene Modal
BGIBMGA007813
Annotation
PREDICTED:_small_integral_membrane_protein_8_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.184 Mitochondrial Reliability : 1.613 PlasmaMembrane Reliability : 1.084
Sequence
CDS
ATGTCAAAACCACACGATGATAAACCTGGCGAGGGTTTGCGATCTATGCGATCAACTACAGCATTTAGAGTGGTCAATTTTGAACTATACGCAAAACCCAATATAGTCATAATGAGCATAGGACTGACATGCTTTGGTTTGGCTCTAGGATACATTGCATACATGCGACAAAAATATGAATCAATGGGATATTATTCTGCTATTGATAAAGATGGAAAAGAGATTTTTGAAAAGAAGAAATCAAAATGGGATTGA
Protein
MSKPHDDKPGEGLRSMRSTTAFRVVNFELYAKPNIVIMSIGLTCFGLALGYIAYMRQKYESMGYYSAIDKDGKEIFEKKKSKWD
Summary
Uniprot
H9JE66
A0A0N1IGM0
I4DL51
S4PFU2
A0A3S2LC47
A0A2H1VMK0
+ More
A0A212F335 A0A182RS38 A0A182JNU0 A0A182Q7Q7 A0A182VWT7 A0A182J4S5 A0A084VXX1 A0A182NRP6 A0A182YHN7 A0A182V6A5 A0A182LEW3 A0A182XHV0 Q5TTT9 A0A182P958 A0A2M3ZCK1 A0A2M4AUH5 A0A2M4C4U2 W5J4W3 A0A182FRJ4 A0A182HST3 A0A2J7Q7M6 A0A1L8D9N1 A0A067RL53 A0A023ECM2 Q16ZB5 A0A023EEL0 A0A182GJB6 A0A023EEL5 A0A336MQS8 A0A336KE03 U5EFY7 A0A1Q3FYK2 D6WMW2 A0A1J1IKQ1 B3MA46 A0A1A9WG94 B4J0R6 A0A2P8XEP6 B4PI39 Q2M040 B4GS56 B4LC06 B4MLY6 A0A023ECJ6 B3NJ03 A0A3B0JRT9 A0A1L8E8K8 A0A1B0BYU2 B4KZS6 N6UJ66 B4IAW5 B4QKN9 Q8IPS9 A0A1W4UE42 A0A1B0GD52 B0X0T2 A0A0L0CQR9 A0A0M4F216 A0A1I8N7K2 A0A1I8NVZ1 A0A1I8NW00 T1GFP4 A0A232F6X4 K7J4Q4 E0VJ51 J9K235 A0A1W4XBJ1 E9HGM8 A0A162QQG9 A0A0L7QQR1 A0A0N7ZHL7 A0A444SER9 A0A0M9ABK3 A0A0P5FF72 A0A0P5SS35 A0A1W2WIF6 A0A1D2NMR9 A0A1S3DPJ1 T1JP98 A0A026VXY0 A0A210PGS0 A8E705 A0A2C9K070 A0A151IG85 A0A088ASK0 A0A1S3GI01 E9JCE8 K9IGI0 I3MKJ7 A0A3P9I130 A0A195FLF2 Q4SEM7
A0A212F335 A0A182RS38 A0A182JNU0 A0A182Q7Q7 A0A182VWT7 A0A182J4S5 A0A084VXX1 A0A182NRP6 A0A182YHN7 A0A182V6A5 A0A182LEW3 A0A182XHV0 Q5TTT9 A0A182P958 A0A2M3ZCK1 A0A2M4AUH5 A0A2M4C4U2 W5J4W3 A0A182FRJ4 A0A182HST3 A0A2J7Q7M6 A0A1L8D9N1 A0A067RL53 A0A023ECM2 Q16ZB5 A0A023EEL0 A0A182GJB6 A0A023EEL5 A0A336MQS8 A0A336KE03 U5EFY7 A0A1Q3FYK2 D6WMW2 A0A1J1IKQ1 B3MA46 A0A1A9WG94 B4J0R6 A0A2P8XEP6 B4PI39 Q2M040 B4GS56 B4LC06 B4MLY6 A0A023ECJ6 B3NJ03 A0A3B0JRT9 A0A1L8E8K8 A0A1B0BYU2 B4KZS6 N6UJ66 B4IAW5 B4QKN9 Q8IPS9 A0A1W4UE42 A0A1B0GD52 B0X0T2 A0A0L0CQR9 A0A0M4F216 A0A1I8N7K2 A0A1I8NVZ1 A0A1I8NW00 T1GFP4 A0A232F6X4 K7J4Q4 E0VJ51 J9K235 A0A1W4XBJ1 E9HGM8 A0A162QQG9 A0A0L7QQR1 A0A0N7ZHL7 A0A444SER9 A0A0M9ABK3 A0A0P5FF72 A0A0P5SS35 A0A1W2WIF6 A0A1D2NMR9 A0A1S3DPJ1 T1JP98 A0A026VXY0 A0A210PGS0 A8E705 A0A2C9K070 A0A151IG85 A0A088ASK0 A0A1S3GI01 E9JCE8 K9IGI0 I3MKJ7 A0A3P9I130 A0A195FLF2 Q4SEM7
Pubmed
19121390
26354079
22651552
23622113
22118469
24438588
+ More
25244985 20966253 12364791 14747013 17210077 20920257 23761445 24845553 24945155 17510324 26483478 18362917 19820115 17994087 29403074 17550304 15632085 23537049 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 25315136 28648823 20075255 20566863 21292972 27289101 24508170 30249741 28812685 15562597 21282665 17554307 15496914
25244985 20966253 12364791 14747013 17210077 20920257 23761445 24845553 24945155 17510324 26483478 18362917 19820115 17994087 29403074 17550304 15632085 23537049 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 25315136 28648823 20075255 20566863 21292972 27289101 24508170 30249741 28812685 15562597 21282665 17554307 15496914
EMBL
BABH01002645
KQ459937
KPJ19231.1
AK402019
KQ459602
BAM18641.1
+ More
KPI93278.1 GAIX01003882 JAA88678.1 RSAL01000049 RVE50398.1 ODYU01003218 SOQ41672.1 AGBW02010631 OWR48144.1 AXCN02000007 ATLV01018209 KE525224 KFB42815.1 AAAB01008859 EAL40900.3 GGFM01005536 MBW26287.1 GGFK01011125 MBW44446.1 GGFJ01011235 MBW60376.1 ADMH02002105 ETN58991.1 APCN01000347 NEVH01017441 PNF24586.1 GFDF01010906 JAV03178.1 KK852561 KDR21335.1 GAPW01006496 JAC07102.1 CH477494 EAT39987.1 EAT39988.1 EAT39989.1 EAT39990.1 GAPW01006493 JAC07105.1 JXUM01067752 KQ562466 KXJ75845.1 GAPW01006494 JAC07104.1 UFQT01002149 SSX32772.1 UFQS01000305 UFQT01000305 SSX02681.1 SSX23055.1 GANO01003678 JAB56193.1 GFDL01002422 JAV32623.1 KQ971343 EFA03274.1 CVRI01000048 CRK99033.1 CH902618 EDV41266.1 CH916366 EDV97921.1 PYGN01002443 PSN30473.1 CM000159 EDW95487.1 CH379069 EAL31096.2 CH479188 EDW40591.1 CH940647 EDW69806.1 CH963847 EDW73197.2 GAPW01006521 JAC07077.1 CH954178 EDV52720.1 OUUW01000002 SPP76419.1 GFDG01003887 JAV14912.1 JXJN01022835 CH933809 EDW19032.1 APGK01032711 KB740848 KB632411 ENN78672.1 ERL95227.1 CH480826 EDW44428.1 CM000363 CM002912 EDX11454.1 KMZ01104.1 AE014296 AAN12189.2 ADV37593.1 CCAG010002317 DS232245 EDS38324.1 JRES01000034 KNC34720.1 CP012525 ALC45095.1 CAQQ02093285 CAQQ02093286 NNAY01000855 OXU26189.1 AAZX01008388 DS235219 EEB13407.1 ABLF02011763 GL732642 EFX69126.1 LRGB01000311 KZS19871.1 KQ414785 KOC60968.1 GDIP01244601 JAI78800.1 SAUD01039730 SAUD01001174 RXG51901.1 RXG72040.1 KQ435710 KOX79913.1 GDIQ01258857 JAJ92867.1 GDIP01136113 JAL67601.1 LJIJ01000003 ODN06527.1 JH431959 KK107796 QOIP01000013 EZA47714.1 RLU15566.1 NEDP02076714 OWF35670.1 BT030947 ABV82329.1 KQ977739 KYN00222.1 GL771805 EFZ09505.1 GABZ01008658 JAA44867.1 AGTP01035943 KQ981512 KYN40819.1 CAAE01014615 CAG00905.1
KPI93278.1 GAIX01003882 JAA88678.1 RSAL01000049 RVE50398.1 ODYU01003218 SOQ41672.1 AGBW02010631 OWR48144.1 AXCN02000007 ATLV01018209 KE525224 KFB42815.1 AAAB01008859 EAL40900.3 GGFM01005536 MBW26287.1 GGFK01011125 MBW44446.1 GGFJ01011235 MBW60376.1 ADMH02002105 ETN58991.1 APCN01000347 NEVH01017441 PNF24586.1 GFDF01010906 JAV03178.1 KK852561 KDR21335.1 GAPW01006496 JAC07102.1 CH477494 EAT39987.1 EAT39988.1 EAT39989.1 EAT39990.1 GAPW01006493 JAC07105.1 JXUM01067752 KQ562466 KXJ75845.1 GAPW01006494 JAC07104.1 UFQT01002149 SSX32772.1 UFQS01000305 UFQT01000305 SSX02681.1 SSX23055.1 GANO01003678 JAB56193.1 GFDL01002422 JAV32623.1 KQ971343 EFA03274.1 CVRI01000048 CRK99033.1 CH902618 EDV41266.1 CH916366 EDV97921.1 PYGN01002443 PSN30473.1 CM000159 EDW95487.1 CH379069 EAL31096.2 CH479188 EDW40591.1 CH940647 EDW69806.1 CH963847 EDW73197.2 GAPW01006521 JAC07077.1 CH954178 EDV52720.1 OUUW01000002 SPP76419.1 GFDG01003887 JAV14912.1 JXJN01022835 CH933809 EDW19032.1 APGK01032711 KB740848 KB632411 ENN78672.1 ERL95227.1 CH480826 EDW44428.1 CM000363 CM002912 EDX11454.1 KMZ01104.1 AE014296 AAN12189.2 ADV37593.1 CCAG010002317 DS232245 EDS38324.1 JRES01000034 KNC34720.1 CP012525 ALC45095.1 CAQQ02093285 CAQQ02093286 NNAY01000855 OXU26189.1 AAZX01008388 DS235219 EEB13407.1 ABLF02011763 GL732642 EFX69126.1 LRGB01000311 KZS19871.1 KQ414785 KOC60968.1 GDIP01244601 JAI78800.1 SAUD01039730 SAUD01001174 RXG51901.1 RXG72040.1 KQ435710 KOX79913.1 GDIQ01258857 JAJ92867.1 GDIP01136113 JAL67601.1 LJIJ01000003 ODN06527.1 JH431959 KK107796 QOIP01000013 EZA47714.1 RLU15566.1 NEDP02076714 OWF35670.1 BT030947 ABV82329.1 KQ977739 KYN00222.1 GL771805 EFZ09505.1 GABZ01008658 JAA44867.1 AGTP01035943 KQ981512 KYN40819.1 CAAE01014615 CAG00905.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000075900
+ More
UP000075881 UP000075886 UP000075920 UP000075880 UP000030765 UP000075884 UP000076408 UP000075903 UP000075882 UP000076407 UP000007062 UP000075885 UP000000673 UP000069272 UP000075840 UP000235965 UP000027135 UP000008820 UP000069940 UP000249989 UP000007266 UP000183832 UP000007801 UP000091820 UP000001070 UP000245037 UP000002282 UP000001819 UP000008744 UP000008792 UP000007798 UP000008711 UP000268350 UP000092460 UP000009192 UP000019118 UP000030742 UP000001292 UP000000304 UP000000803 UP000192221 UP000092444 UP000002320 UP000037069 UP000092553 UP000095301 UP000095300 UP000015102 UP000215335 UP000002358 UP000009046 UP000007819 UP000192223 UP000000305 UP000076858 UP000053825 UP000288706 UP000053105 UP000094527 UP000079169 UP000053097 UP000279307 UP000242188 UP000076420 UP000078542 UP000005203 UP000081671 UP000005215 UP000265200 UP000078541
UP000075881 UP000075886 UP000075920 UP000075880 UP000030765 UP000075884 UP000076408 UP000075903 UP000075882 UP000076407 UP000007062 UP000075885 UP000000673 UP000069272 UP000075840 UP000235965 UP000027135 UP000008820 UP000069940 UP000249989 UP000007266 UP000183832 UP000007801 UP000091820 UP000001070 UP000245037 UP000002282 UP000001819 UP000008744 UP000008792 UP000007798 UP000008711 UP000268350 UP000092460 UP000009192 UP000019118 UP000030742 UP000001292 UP000000304 UP000000803 UP000192221 UP000092444 UP000002320 UP000037069 UP000092553 UP000095301 UP000095300 UP000015102 UP000215335 UP000002358 UP000009046 UP000007819 UP000192223 UP000000305 UP000076858 UP000053825 UP000288706 UP000053105 UP000094527 UP000079169 UP000053097 UP000279307 UP000242188 UP000076420 UP000078542 UP000005203 UP000081671 UP000005215 UP000265200 UP000078541
Pfam
PF14937 DUF4500
Interpro
IPR026686
UPF0708
ProteinModelPortal
H9JE66
A0A0N1IGM0
I4DL51
S4PFU2
A0A3S2LC47
A0A2H1VMK0
+ More
A0A212F335 A0A182RS38 A0A182JNU0 A0A182Q7Q7 A0A182VWT7 A0A182J4S5 A0A084VXX1 A0A182NRP6 A0A182YHN7 A0A182V6A5 A0A182LEW3 A0A182XHV0 Q5TTT9 A0A182P958 A0A2M3ZCK1 A0A2M4AUH5 A0A2M4C4U2 W5J4W3 A0A182FRJ4 A0A182HST3 A0A2J7Q7M6 A0A1L8D9N1 A0A067RL53 A0A023ECM2 Q16ZB5 A0A023EEL0 A0A182GJB6 A0A023EEL5 A0A336MQS8 A0A336KE03 U5EFY7 A0A1Q3FYK2 D6WMW2 A0A1J1IKQ1 B3MA46 A0A1A9WG94 B4J0R6 A0A2P8XEP6 B4PI39 Q2M040 B4GS56 B4LC06 B4MLY6 A0A023ECJ6 B3NJ03 A0A3B0JRT9 A0A1L8E8K8 A0A1B0BYU2 B4KZS6 N6UJ66 B4IAW5 B4QKN9 Q8IPS9 A0A1W4UE42 A0A1B0GD52 B0X0T2 A0A0L0CQR9 A0A0M4F216 A0A1I8N7K2 A0A1I8NVZ1 A0A1I8NW00 T1GFP4 A0A232F6X4 K7J4Q4 E0VJ51 J9K235 A0A1W4XBJ1 E9HGM8 A0A162QQG9 A0A0L7QQR1 A0A0N7ZHL7 A0A444SER9 A0A0M9ABK3 A0A0P5FF72 A0A0P5SS35 A0A1W2WIF6 A0A1D2NMR9 A0A1S3DPJ1 T1JP98 A0A026VXY0 A0A210PGS0 A8E705 A0A2C9K070 A0A151IG85 A0A088ASK0 A0A1S3GI01 E9JCE8 K9IGI0 I3MKJ7 A0A3P9I130 A0A195FLF2 Q4SEM7
A0A212F335 A0A182RS38 A0A182JNU0 A0A182Q7Q7 A0A182VWT7 A0A182J4S5 A0A084VXX1 A0A182NRP6 A0A182YHN7 A0A182V6A5 A0A182LEW3 A0A182XHV0 Q5TTT9 A0A182P958 A0A2M3ZCK1 A0A2M4AUH5 A0A2M4C4U2 W5J4W3 A0A182FRJ4 A0A182HST3 A0A2J7Q7M6 A0A1L8D9N1 A0A067RL53 A0A023ECM2 Q16ZB5 A0A023EEL0 A0A182GJB6 A0A023EEL5 A0A336MQS8 A0A336KE03 U5EFY7 A0A1Q3FYK2 D6WMW2 A0A1J1IKQ1 B3MA46 A0A1A9WG94 B4J0R6 A0A2P8XEP6 B4PI39 Q2M040 B4GS56 B4LC06 B4MLY6 A0A023ECJ6 B3NJ03 A0A3B0JRT9 A0A1L8E8K8 A0A1B0BYU2 B4KZS6 N6UJ66 B4IAW5 B4QKN9 Q8IPS9 A0A1W4UE42 A0A1B0GD52 B0X0T2 A0A0L0CQR9 A0A0M4F216 A0A1I8N7K2 A0A1I8NVZ1 A0A1I8NW00 T1GFP4 A0A232F6X4 K7J4Q4 E0VJ51 J9K235 A0A1W4XBJ1 E9HGM8 A0A162QQG9 A0A0L7QQR1 A0A0N7ZHL7 A0A444SER9 A0A0M9ABK3 A0A0P5FF72 A0A0P5SS35 A0A1W2WIF6 A0A1D2NMR9 A0A1S3DPJ1 T1JP98 A0A026VXY0 A0A210PGS0 A8E705 A0A2C9K070 A0A151IG85 A0A088ASK0 A0A1S3GI01 E9JCE8 K9IGI0 I3MKJ7 A0A3P9I130 A0A195FLF2 Q4SEM7
Ontologies
GO
PANTHER
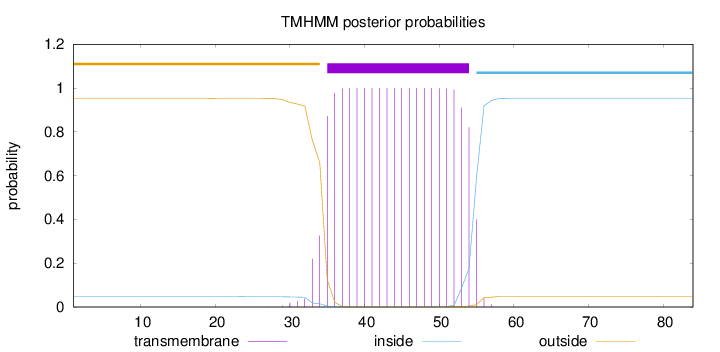
Topology
Length:
84
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.65804
Exp number, first 60 AAs:
20.65804
Total prob of N-in:
0.04735
POSSIBLE N-term signal
sequence
outside
1 - 34
TMhelix
35 - 54
inside
55 - 84
Population Genetic Test Statistics
Pi
153.526043
Theta
205.475124
Tajima's D
-0.701851
CLR
0.24931
CSRT
0.196190190490475
Interpretation
Uncertain
