Gene
KWMTBOMO08921
Pre Gene Modal
BGIBMGA007814
Annotation
PREDICTED:_rab-like_protein_6_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.636
Sequence
CDS
ATGTTTTCTGCTTTGAAAAAGTTAGCTAGAAGCAGTGATGATCGGTGCCCTCCTCCAATGCCAATGTCTTCCTCGTTGCAGAAAAAGTTTTCTAGAGGAGTGCATTTTAATATGAAAATATTGATCAAAGGAGATAGAAATGTAGGTAAATCGTGCCTTCTTCAACGTCTTCAAGGCGGTCCATTTATCGAAGACTATGTTCCAACAGATCAAATACAAGTGGCTCCAATACACTGGACTTATAAAAACAGCGATTATATAGTGAAGGTAGAAGTTTGGGAGGTTGTGGATAAAGGACGTACAAAAAAGAAAACCCCAATAGGACTGAAGTTAGAAAATCAATCAGCCCCATCATTAGAGGACAATCTCGATGCTCCTGTACTAGATGCAACCTTTTTGGATGTCTACAAAAATACCACTGGTGTTATACTTATGCTTGATATAACTAAGCCTTGGACATTTGAATATGTTGTGAAAGAACTATCACAAGTACCTGCAGATTTGCCAGTTGTTATCTTAGGAAACCATTGTGATATGCAGCACCATCGCCAAGTGCATTCCCACCATATTGAACAGGCTTTACATAATGCAAAGCAAATTAGAACAGCTCCAGTTCGTTATGCAGAATCGTCCATGAGAAATGGCTTTGGATTGCGTTTATTACATAAGTTTCTGAGTATACCATTTTTGCGCCTTCAAAAGACAAGTTTGTTAGAGCAACTACAGAGAAATCAAAGAGATATGGAAGAAATAGAGCAGGAATTAGATGAATATCAGAATTCAGAAGAATCACAGTACAACACATTCCTAGATCGTTTAACAAATAAAAGGAAACAGAACACTAATCAAGTTGAACCTCGACCAAGCAATGAACGCTCTCCTAGCATTGTACTAGGTGGTGGGAAACCCATAGTGCCACCCAGTGTAAATCCATTACTGGCCCAATCTCTAAATCCATCAATGAAAGCACAAATACCAGAAGCTAAGCAAACTCAATTTGTTCATCCTGATCTTATAAAACCTAAGCAAACTGTAAACATGGACACGACGCCGCCACGCCTCCTCCAACACGATAGTAATCAAAGCACACCTTCAGTCACAAATGCGGGTAGTTCTGTTGGAACTCTAGACGAATTTTATGCTGGTGCACTAGATAGTAGCTTCTTGGAGGATCTAACACCTTTGTCTAGTAGTAGACAGGAGATACAATTTGAATCGGAAAGTGATGATGACACTACAAATCCAAAAGTTACAGTAGATGAAGATGATCTTGATATCGATACTTTTTCGACTGAATTGACACAAAAAACAAAGCTGCAACAAAAAGACCAGCTAGTGGAAACAGATTTACATAAAACGATTGAAAGAAATACAAGCCAAAAGCGAGAGAGTGAAACGGAATTTGAAAAGACATCAAATCCAGATATAAATTTACCTATATTTGAAAATAGACATAACATCCAATTGTCTTTGGATAATGACTTCCCTCTATGGAGTGGTGACTCTAGCGTTAGAAGATCACCTGAAGGTGGCGAAGATCCTGATGCCCTTAAAGATCCTGGTGATAAGTCTGAAAAAAAACACCACAAGAAGAAAAAGTCCAAAGATAAAGACCGCGGCGACTACAGTGAGATATCAGAACGCAAAGAGAAGAAACAAAAACGTAAGAAGTCTAAAAGTGAAAAAAAACCCAATCCTCAGCAGGACTTGTTAGCGCCTTCAGGATCTGTAGACTTTGACTATGCAGAATATGATAGCATTTAA
Protein
MFSALKKLARSSDDRCPPPMPMSSSLQKKFSRGVHFNMKILIKGDRNVGKSCLLQRLQGGPFIEDYVPTDQIQVAPIHWTYKNSDYIVKVEVWEVVDKGRTKKKTPIGLKLENQSAPSLEDNLDAPVLDATFLDVYKNTTGVILMLDITKPWTFEYVVKELSQVPADLPVVILGNHCDMQHHRQVHSHHIEQALHNAKQIRTAPVRYAESSMRNGFGLRLLHKFLSIPFLRLQKTSLLEQLQRNQRDMEEIEQELDEYQNSEESQYNTFLDRLTNKRKQNTNQVEPRPSNERSPSIVLGGGKPIVPPSVNPLLAQSLNPSMKAQIPEAKQTQFVHPDLIKPKQTVNMDTTPPRLLQHDSNQSTPSVTNAGSSVGTLDEFYAGALDSSFLEDLTPLSSSRQEIQFESESDDDTTNPKVTVDEDDLDIDTFSTELTQKTKLQQKDQLVETDLHKTIERNTSQKRESETEFEKTSNPDINLPIFENRHNIQLSLDNDFPLWSGDSSVRRSPEGGEDPDALKDPGDKSEKKHHKKKKSKDKDRGDYSEISERKEKKQKRKKSKSEKKPNPQQDLLAPSGSVDFDYAEYDSI
Summary
Uniprot
H9JE67
A0A1E1WF33
A0A194PIY0
A0A0N1PJA1
A0A2H1VLI4
A0A3S2P245
+ More
A0A0L7L6X4 A0A212EWS9 U5EVA4 A0A1W4XIH6 A0A1S4G7J3 B0WEP3 D7EI68 A0A1Q3FXS6 A0A2A3EFL4 A0A336KN38 A0A088A5F6 A0A182JTI7 A0A154PE97 A0A1Y1MJX3 A0A182S612 A0A182QYT9 A0A0L7R5C3 A0A182Y1M7 A0A182RKT4 A0A182WG53 A0A0N0BJG7 A0A182TIF1 A0A232EV50 A0A182HQ82 A0A182UPY2 A0A182MYQ2 A0A182KSG3 A0A1S4HF81 V5I937 A0A182WZN6 A0A182P6J9 N6TWW8 A0A1B6CF37 A0A1L8DLN8 A0A2R5LBX7 A0A182MKJ9 A0A084VI87 E0VGD9 T1K7M9 E2AA22 A0A182H3G5 A0A1B0D020 A0A182FTU3 A0A2M4BHH7 A0A0C9S2Q0 F4WND2 A0A151XAN4 W5JM37 A0A2S2PV14 A0A158NG81 A0A195B7Y2 A0A195FF72 A0A151J5C5 A0A0V0G5B7 A0A1B6MKQ6 A0A0J7KX05 A0A195CK01 E2BXL8 A0A1L8F5Q8 A0A026W380 A0A1L8F0V6 A0A0P4VVJ9 A0A224XNA6 A0A069DZ75 A0A3L8DMG8
A0A0L7L6X4 A0A212EWS9 U5EVA4 A0A1W4XIH6 A0A1S4G7J3 B0WEP3 D7EI68 A0A1Q3FXS6 A0A2A3EFL4 A0A336KN38 A0A088A5F6 A0A182JTI7 A0A154PE97 A0A1Y1MJX3 A0A182S612 A0A182QYT9 A0A0L7R5C3 A0A182Y1M7 A0A182RKT4 A0A182WG53 A0A0N0BJG7 A0A182TIF1 A0A232EV50 A0A182HQ82 A0A182UPY2 A0A182MYQ2 A0A182KSG3 A0A1S4HF81 V5I937 A0A182WZN6 A0A182P6J9 N6TWW8 A0A1B6CF37 A0A1L8DLN8 A0A2R5LBX7 A0A182MKJ9 A0A084VI87 E0VGD9 T1K7M9 E2AA22 A0A182H3G5 A0A1B0D020 A0A182FTU3 A0A2M4BHH7 A0A0C9S2Q0 F4WND2 A0A151XAN4 W5JM37 A0A2S2PV14 A0A158NG81 A0A195B7Y2 A0A195FF72 A0A151J5C5 A0A0V0G5B7 A0A1B6MKQ6 A0A0J7KX05 A0A195CK01 E2BXL8 A0A1L8F5Q8 A0A026W380 A0A1L8F0V6 A0A0P4VVJ9 A0A224XNA6 A0A069DZ75 A0A3L8DMG8
Pubmed
EMBL
BABH01002645
GDQN01005454
JAT85600.1
KQ459602
KPI93277.1
KQ459937
+ More
KPJ19230.1 ODYU01003218 SOQ41671.1 RSAL01000049 RVE50401.1 JTDY01002537 KOB71223.1 AGBW02011886 OWR45968.1 GANO01001945 JAB57926.1 DS231910 EDS45759.1 DS497673 EFA13241.1 GFDL01002752 JAV32293.1 KZ288256 PBC30545.1 UFQS01000691 UFQT01000691 SSX06246.1 SSX26600.1 KQ434874 KZC09744.1 GEZM01029626 JAV86033.1 AXCN02001032 KQ414654 KOC65961.1 KQ435715 KOX79088.1 NNAY01002053 OXU22229.1 APCN01003297 AAAB01008986 GALX01003615 JAB64851.1 APGK01048477 KB741112 ENN73795.1 GEDC01025285 JAS12013.1 GFDF01006708 JAV07376.1 GGLE01002915 MBY07041.1 AXCM01007497 ATLV01013330 KE524853 KFB37681.1 DS235138 EEB12445.1 CAEY01001807 GL438001 EFN69721.1 JXUM01107438 KQ565141 KXJ71290.1 AJVK01021173 GGFJ01003333 MBW52474.1 GBYB01012299 GBYB01015181 JAG82066.1 JAG84948.1 GL888236 EGI64302.1 KQ982339 KYQ57389.1 ADMH02000662 ETN65412.1 GGMR01020077 MBY32696.1 ADTU01015041 KQ976574 KYM80352.1 KQ981625 KYN39011.1 KQ980017 KYN18234.1 GECL01002857 JAP03267.1 GEBQ01003483 JAT36494.1 LBMM01002422 KMQ94846.1 KQ977721 KYN00399.1 GL451276 EFN79562.1 CM004480 OCT66919.1 KK107487 EZA50041.1 CM004481 OCT65208.1 GDKW01000519 JAI56076.1 GFTR01006957 JAW09469.1 GBGD01001130 JAC87759.1 QOIP01000006 RLU21119.1
KPJ19230.1 ODYU01003218 SOQ41671.1 RSAL01000049 RVE50401.1 JTDY01002537 KOB71223.1 AGBW02011886 OWR45968.1 GANO01001945 JAB57926.1 DS231910 EDS45759.1 DS497673 EFA13241.1 GFDL01002752 JAV32293.1 KZ288256 PBC30545.1 UFQS01000691 UFQT01000691 SSX06246.1 SSX26600.1 KQ434874 KZC09744.1 GEZM01029626 JAV86033.1 AXCN02001032 KQ414654 KOC65961.1 KQ435715 KOX79088.1 NNAY01002053 OXU22229.1 APCN01003297 AAAB01008986 GALX01003615 JAB64851.1 APGK01048477 KB741112 ENN73795.1 GEDC01025285 JAS12013.1 GFDF01006708 JAV07376.1 GGLE01002915 MBY07041.1 AXCM01007497 ATLV01013330 KE524853 KFB37681.1 DS235138 EEB12445.1 CAEY01001807 GL438001 EFN69721.1 JXUM01107438 KQ565141 KXJ71290.1 AJVK01021173 GGFJ01003333 MBW52474.1 GBYB01012299 GBYB01015181 JAG82066.1 JAG84948.1 GL888236 EGI64302.1 KQ982339 KYQ57389.1 ADMH02000662 ETN65412.1 GGMR01020077 MBY32696.1 ADTU01015041 KQ976574 KYM80352.1 KQ981625 KYN39011.1 KQ980017 KYN18234.1 GECL01002857 JAP03267.1 GEBQ01003483 JAT36494.1 LBMM01002422 KMQ94846.1 KQ977721 KYN00399.1 GL451276 EFN79562.1 CM004480 OCT66919.1 KK107487 EZA50041.1 CM004481 OCT65208.1 GDKW01000519 JAI56076.1 GFTR01006957 JAW09469.1 GBGD01001130 JAC87759.1 QOIP01000006 RLU21119.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000037510
UP000007151
+ More
UP000192223 UP000002320 UP000007266 UP000242457 UP000005203 UP000075881 UP000076502 UP000075901 UP000075886 UP000053825 UP000076408 UP000075900 UP000075920 UP000053105 UP000075902 UP000215335 UP000075840 UP000075903 UP000075884 UP000075882 UP000076407 UP000075885 UP000019118 UP000075883 UP000030765 UP000009046 UP000015104 UP000000311 UP000069940 UP000249989 UP000092462 UP000069272 UP000007755 UP000075809 UP000000673 UP000005205 UP000078540 UP000078541 UP000078492 UP000036403 UP000078542 UP000008237 UP000186698 UP000053097 UP000279307
UP000192223 UP000002320 UP000007266 UP000242457 UP000005203 UP000075881 UP000076502 UP000075901 UP000075886 UP000053825 UP000076408 UP000075900 UP000075920 UP000053105 UP000075902 UP000215335 UP000075840 UP000075903 UP000075884 UP000075882 UP000076407 UP000075885 UP000019118 UP000075883 UP000030765 UP000009046 UP000015104 UP000000311 UP000069940 UP000249989 UP000092462 UP000069272 UP000007755 UP000075809 UP000000673 UP000005205 UP000078540 UP000078541 UP000078492 UP000036403 UP000078542 UP000008237 UP000186698 UP000053097 UP000279307
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JE67
A0A1E1WF33
A0A194PIY0
A0A0N1PJA1
A0A2H1VLI4
A0A3S2P245
+ More
A0A0L7L6X4 A0A212EWS9 U5EVA4 A0A1W4XIH6 A0A1S4G7J3 B0WEP3 D7EI68 A0A1Q3FXS6 A0A2A3EFL4 A0A336KN38 A0A088A5F6 A0A182JTI7 A0A154PE97 A0A1Y1MJX3 A0A182S612 A0A182QYT9 A0A0L7R5C3 A0A182Y1M7 A0A182RKT4 A0A182WG53 A0A0N0BJG7 A0A182TIF1 A0A232EV50 A0A182HQ82 A0A182UPY2 A0A182MYQ2 A0A182KSG3 A0A1S4HF81 V5I937 A0A182WZN6 A0A182P6J9 N6TWW8 A0A1B6CF37 A0A1L8DLN8 A0A2R5LBX7 A0A182MKJ9 A0A084VI87 E0VGD9 T1K7M9 E2AA22 A0A182H3G5 A0A1B0D020 A0A182FTU3 A0A2M4BHH7 A0A0C9S2Q0 F4WND2 A0A151XAN4 W5JM37 A0A2S2PV14 A0A158NG81 A0A195B7Y2 A0A195FF72 A0A151J5C5 A0A0V0G5B7 A0A1B6MKQ6 A0A0J7KX05 A0A195CK01 E2BXL8 A0A1L8F5Q8 A0A026W380 A0A1L8F0V6 A0A0P4VVJ9 A0A224XNA6 A0A069DZ75 A0A3L8DMG8
A0A0L7L6X4 A0A212EWS9 U5EVA4 A0A1W4XIH6 A0A1S4G7J3 B0WEP3 D7EI68 A0A1Q3FXS6 A0A2A3EFL4 A0A336KN38 A0A088A5F6 A0A182JTI7 A0A154PE97 A0A1Y1MJX3 A0A182S612 A0A182QYT9 A0A0L7R5C3 A0A182Y1M7 A0A182RKT4 A0A182WG53 A0A0N0BJG7 A0A182TIF1 A0A232EV50 A0A182HQ82 A0A182UPY2 A0A182MYQ2 A0A182KSG3 A0A1S4HF81 V5I937 A0A182WZN6 A0A182P6J9 N6TWW8 A0A1B6CF37 A0A1L8DLN8 A0A2R5LBX7 A0A182MKJ9 A0A084VI87 E0VGD9 T1K7M9 E2AA22 A0A182H3G5 A0A1B0D020 A0A182FTU3 A0A2M4BHH7 A0A0C9S2Q0 F4WND2 A0A151XAN4 W5JM37 A0A2S2PV14 A0A158NG81 A0A195B7Y2 A0A195FF72 A0A151J5C5 A0A0V0G5B7 A0A1B6MKQ6 A0A0J7KX05 A0A195CK01 E2BXL8 A0A1L8F5Q8 A0A026W380 A0A1L8F0V6 A0A0P4VVJ9 A0A224XNA6 A0A069DZ75 A0A3L8DMG8
PDB
6O62
E-value=1.59977e-05,
Score=117
Ontologies
GO
PANTHER
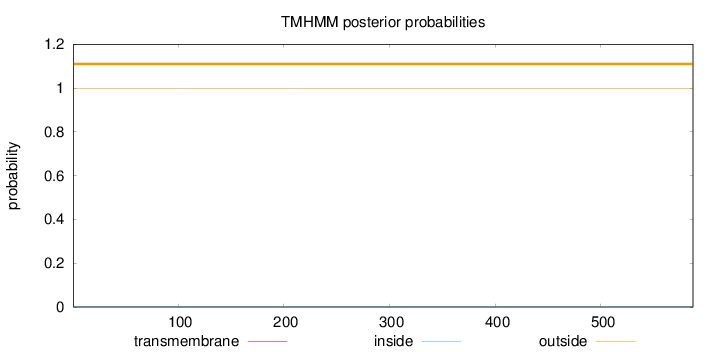
Topology
Length:
587
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00142
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00074
outside
1 - 587
Population Genetic Test Statistics
Pi
201.702578
Theta
190.168157
Tajima's D
0.34859
CLR
0.048964
CSRT
0.467276636168192
Interpretation
Uncertain
