Gene
KWMTBOMO08919
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.15
Sequence
CDS
ATGCGCAAACGCCAACACTGGATGACCGATGGTACGCTCGCCCTAATTGAAGAGAGACGTCGAAAGAAAGCTCAAGGCATAGACACGAACAAACTGAATGTATTGTCTGCCAATATTCAGGCCAGCTGCAGACGAGATCATAACTCTCATCTTCGTAACATTTGTGAAGAGGTCGAAAAACATGCTATGAAGCATGAATCTAGAGATCTCTACCACAAAATACGGTTCATTACCAAATCACTGCCATCTAAGACCTGGGCTATTGTAAATGATAAAAACGAACTGATAACAGAGTTGGATCAAATCTCTGAAACATGGAAGAGTTACTGTCAATCGTTATTCCAGGATCCACAGTCACGGCAATTTACAAGTACAGACCCAAACGATGAGGACTTGGAGCCAAGCATTCTCCTGTCTGAGGTTAGGGCTGCTATAAAACACCTCAAAAACAGGAAAGCGACTGGTAGAGATGCAATTCCTATTGAAACAATAAAAGCCTTAGGAGAACGCGGTGACCACATATTTCATAAGATATGCAACAGGGTATGGCAGACGGGAGTTTGGCCGTCGGAATGGACACATACCGTGTTTACTCCCCTGCATAAAAAGGGCTCTACGAAGAAATGTAACAACTATCGTCTAATTGCTCTTACATCACATTCTAGCAAAATTATGTTACATATTCTGAATGAGCGATTAAAAACCTATCTGTCCAAAGAGATTGCTCCGGAACAGGCCGGTTTCGTGAGGGGGAAAGGCACTCGGGAACAAATTTTCATCGTACGTCAGATAATTGAGAAAGCAAGAGAATTCAACAGGCCGACATACATTTGTTTTGTTGACTTCTCCAAAGCATTTGACTCGGTGAAATGGCCTGTTTTGTGGAGGACCTTACTGGATCTGGGGACACCGAAACATCTTGTGCACCTACTGAGACGCTTATATGAGAATGGTACGGCCTCGGTGCGCGCGGATGACGTTCTCTCTGATAACTTCCATCCAAGTGCTGGGGTTCGCCAAGGTTGCATTGTATCGCCGCTCTTATTCAATGCGTACACAGAAATCATAATGCGCATCACTTTGGAGAACTGGACAGATGGTGTAGCAATTGGGGGGTATAGGATTGCAAACTTGCGATACGCGGATGATACCACTTTGTTTGCAACTGATGCGCAATGCTTGGGGGAGTTACTGTCGAGGATGGAACGTGTGAGCCTTGAGTTTGGATTAAAGATTAACCGCAGTAAGACCAAGGTGATGATTGTCGATCGCGCTATGAATAACTCGCCGGACGTGACTCAAATAGCGGGTTGCGACGTGGTCCAGTCCTACATATACCTTGGGGCGTTAATCTCAAATAATGGCGGATGCATGGACGAAATCAGAAGGCGCATGGCTATCACGAGGTCGGCGATGGACGGGCTGAGGAAAGTATGGAGAAATAGAAACATAACCAAAACCACAAAGATCAGGCTCGTTAGAACACTAATATTCTCTATATTTCTATACGCTGCTGAGACGTGGACCGTGCGAGACGTGGAAAGGAAGAAAATAGACGCTCTGGAAATGTGGTGCTGGAGAAGAATGCTTGGAGTTTCGTGGACCGACTTTCGTACAAATGTCTCGATACTTCAGGAACTCGGCATCAAGCAGCGTCTATTTGGTATAGTACAGTCTCGAATGGTGAATTTCTTCGGACACGTTTCGCGACGAGATGGCCGGTCCATAGAACGCCTCGTTGTACAGGGAAGCGTTGATGGTACAAGACCGCGCGGGAGGCCACCAATGCGGTGGACCGACCAAATTAAAACTGCAGTGGGAGGTCCCCTAAATGAGTGCAGTAGAATGGCCTCGAGCAGGGAGAGATGGCGCGACATCGTGAGACGCATCAAGTCTGCCCCTTCCAACACTACATGA
Protein
MRKRQHWMTDGTLALIEERRRKKAQGIDTNKLNVLSANIQASCRRDHNSHLRNICEEVEKHAMKHESRDLYHKIRFITKSLPSKTWAIVNDKNELITELDQISETWKSYCQSLFQDPQSRQFTSTDPNDEDLEPSILLSEVRAAIKHLKNRKATGRDAIPIETIKALGERGDHIFHKICNRVWQTGVWPSEWTHTVFTPLHKKGSTKKCNNYRLIALTSHSSKIMLHILNERLKTYLSKEIAPEQAGFVRGKGTREQIFIVRQIIEKAREFNRPTYICFVDFSKAFDSVKWPVLWRTLLDLGTPKHLVHLLRRLYENGTASVRADDVLSDNFHPSAGVRQGCIVSPLLFNAYTEIIMRITLENWTDGVAIGGYRIANLRYADDTTLFATDAQCLGELLSRMERVSLEFGLKINRSKTKVMIVDRAMNNSPDVTQIAGCDVVQSYIYLGALISNNGGCMDEIRRRMAITRSAMDGLRKVWRNRNITKTTKIRLVRTLIFSIFLYAAETWTVRDVERKKIDALEMWCWRRMLGVSWTDFRTNVSILQELGIKQRLFGIVQSRMVNFFGHVSRRDGRSIERLVVQGSVDGTRPRGRPPMRWTDQIKTAVGGPLNECSRMASSRERWRDIVRRIKSAPSNTT
Summary
Uniprot
D7F175
D7F167
D7F162
D7F161
D7F163
D7F172
+ More
D7F168 D5LB39 A0A402EB12 A0A402EKC4 A0A2W1BNH4 A0A402EKB1 A0A402EST4 A0A402E768 A0A402EWU2 A0A402E7R7 A0A402EBP1 A0A402F9A7 A0A402ET04 A0A402EWP9 A0A402ECS2 A0A402ET28 A0A402F4E6 A0A402F9C1 A0A402F406 A0A402E7Q0 A0A402F125 A0A402EBC2 A0A402ELM7 A0A402FC20 A0A402FAG4 A0A402F575 A0A402EEU1 A0A402EWN2 A0A402F3F3 A0A402EKM3 A0A402EZW9 A0A402EVG7 A0A402EKK1 A0A402EWI9 A0A402EBI2 A0A402EXG4 A0A443QYR6 A0A402F382 A0A402EWK9 A0A402F9T4 A0A2H6KKM9 A0A402EWI8 A0A402ET45 A0A402EL35 A0A3S3P648 A0A3Q1LQN1 A0A402F7I8 A0A402F3Q6 H3B2Y3 A0A3Q1MLZ9 A0A3Q1M6J7 A0A402ER19 A0A3Q1MJK8 A0A0J7KGF7 A0A3Q1MI63 A0A3Q1MQ02 A0A3Q1M1T7 H2YWZ5 A0A3Q1MMW6 A0A3Q1N3Y9 A0A3Q1MGH6 A0A3Q1LWU3 A0A3Q1MP18 A0A3Q1M8D1 A0A3Q1LR18 A0A3Q1LUA6 A0A3Q1MA23 A0A3Q1LXQ5 A0A3Q1M527 A0A3Q1MDY8 A0A3Q1NE48 A0A3Q1MXT5 A0A3Q1MLI2 A0A402F6U7 A0A3Q1LI58 A0A3Q1N1P1 A0A3Q1N2X2 A0A3Q1MTQ8 A0A3Q1LNZ2 A0A3Q1MZJ0 A0A3Q1LX64 A0A3Q1MWB3 A0A3Q1LXF5 A0A3Q1MLV4 A0A3Q1ML42 A0A3Q1MNB9 A0A3Q1MM54 A0A3Q1LRD2 A0A3Q1LYT3 A0A3Q1LR52
D7F168 D5LB39 A0A402EB12 A0A402EKC4 A0A2W1BNH4 A0A402EKB1 A0A402EST4 A0A402E768 A0A402EWU2 A0A402E7R7 A0A402EBP1 A0A402F9A7 A0A402ET04 A0A402EWP9 A0A402ECS2 A0A402ET28 A0A402F4E6 A0A402F9C1 A0A402F406 A0A402E7Q0 A0A402F125 A0A402EBC2 A0A402ELM7 A0A402FC20 A0A402FAG4 A0A402F575 A0A402EEU1 A0A402EWN2 A0A402F3F3 A0A402EKM3 A0A402EZW9 A0A402EVG7 A0A402EKK1 A0A402EWI9 A0A402EBI2 A0A402EXG4 A0A443QYR6 A0A402F382 A0A402EWK9 A0A402F9T4 A0A2H6KKM9 A0A402EWI8 A0A402ET45 A0A402EL35 A0A3S3P648 A0A3Q1LQN1 A0A402F7I8 A0A402F3Q6 H3B2Y3 A0A3Q1MLZ9 A0A3Q1M6J7 A0A402ER19 A0A3Q1MJK8 A0A0J7KGF7 A0A3Q1MI63 A0A3Q1MQ02 A0A3Q1M1T7 H2YWZ5 A0A3Q1MMW6 A0A3Q1N3Y9 A0A3Q1MGH6 A0A3Q1LWU3 A0A3Q1MP18 A0A3Q1M8D1 A0A3Q1LR18 A0A3Q1LUA6 A0A3Q1MA23 A0A3Q1LXQ5 A0A3Q1M527 A0A3Q1MDY8 A0A3Q1NE48 A0A3Q1MXT5 A0A3Q1MLI2 A0A402F6U7 A0A3Q1LI58 A0A3Q1N1P1 A0A3Q1N2X2 A0A3Q1MTQ8 A0A3Q1LNZ2 A0A3Q1MZJ0 A0A3Q1LX64 A0A3Q1MWB3 A0A3Q1LXF5 A0A3Q1MLV4 A0A3Q1ML42 A0A3Q1MNB9 A0A3Q1MM54 A0A3Q1LRD2 A0A3Q1LYT3 A0A3Q1LR52
EMBL
FJ265560
ADI61828.1
FJ265552
ADI61820.1
FJ265547
ADI61815.1
+ More
FJ265546 ADI61814.1 FJ265548 ADI61816.1 FJ265557 ADI61825.1 FJ265553 ADI61821.1 GU815090 ADF18553.1 BDOT01000005 GCF41419.1 BDOT01000017 GCF44634.1 KZ150025 PZC74827.1 GCF44637.1 BDOT01000038 GCF47275.1 BDOT01000002 GCF39993.1 BDOT01000056 GCF48696.1 GCF40259.1 GCF41584.1 BDOT01000150 GCF53030.1 GCF47332.1 BDOT01000054 GCF48631.1 BDOT01000006 GCF41910.1 BDOT01000039 GCF47373.1 BDOT01000101 GCF51279.1 GCF53027.1 BDOT01000094 GCF51020.1 GCF40210.1 BDOT01000077 GCF50202.1 GCF41464.1 BDOT01000022 GCF45061.1 BDOT01000183 GCF53956.1 BDOT01000165 GCF53431.1 BDOT01000105 GCF51450.1 BDOT01000008 GCF42705.1 GCF48632.1 BDOT01000090 GCF50924.1 BDOT01000018 GCF44752.1 BDOT01000069 GCF49674.1 BDOT01000049 GCF48208.1 GCF44732.1 GCF48586.1 GCF41524.1 BDOT01000059 GCF48922.1 NCKU01003098 RWS08177.1 GCF50913.1 GCF48608.1 BDOT01000156 GCF53186.1 BDSA01000081 GBE63537.1 GCF48559.1 GCF47377.1 BDOT01000020 GCF44897.1 NCKU01004395 RWS06003.1 BDOT01000130 GCF52369.1 GCF51029.1 AFYH01119920 BDOT01000031 GCF46561.1 LBMM01007694 KMQ89503.1 BDOT01000127 GCF52252.1
FJ265546 ADI61814.1 FJ265548 ADI61816.1 FJ265557 ADI61825.1 FJ265553 ADI61821.1 GU815090 ADF18553.1 BDOT01000005 GCF41419.1 BDOT01000017 GCF44634.1 KZ150025 PZC74827.1 GCF44637.1 BDOT01000038 GCF47275.1 BDOT01000002 GCF39993.1 BDOT01000056 GCF48696.1 GCF40259.1 GCF41584.1 BDOT01000150 GCF53030.1 GCF47332.1 BDOT01000054 GCF48631.1 BDOT01000006 GCF41910.1 BDOT01000039 GCF47373.1 BDOT01000101 GCF51279.1 GCF53027.1 BDOT01000094 GCF51020.1 GCF40210.1 BDOT01000077 GCF50202.1 GCF41464.1 BDOT01000022 GCF45061.1 BDOT01000183 GCF53956.1 BDOT01000165 GCF53431.1 BDOT01000105 GCF51450.1 BDOT01000008 GCF42705.1 GCF48632.1 BDOT01000090 GCF50924.1 BDOT01000018 GCF44752.1 BDOT01000069 GCF49674.1 BDOT01000049 GCF48208.1 GCF44732.1 GCF48586.1 GCF41524.1 BDOT01000059 GCF48922.1 NCKU01003098 RWS08177.1 GCF50913.1 GCF48608.1 BDOT01000156 GCF53186.1 BDSA01000081 GBE63537.1 GCF48559.1 GCF47377.1 BDOT01000020 GCF44897.1 NCKU01004395 RWS06003.1 BDOT01000130 GCF52369.1 GCF51029.1 AFYH01119920 BDOT01000031 GCF46561.1 LBMM01007694 KMQ89503.1 BDOT01000127 GCF52252.1
Proteomes
Pfam
Interpro
IPR005135
Endo/exonuclease/phosphatase
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR002138 Pept_C14_p10
IPR035892 C2_domain_sf
IPR000008 C2_dom
IPR030431 FAM189
IPR016024 ARM-type_fold
IPR017956 AT_hook_DNA-bd_motif
IPR039776 Pds5
IPR003349 JmjN
IPR001606 ARID_dom
IPR004198 Znf_C5HC2
IPR036431 ARID_dom_sf
IPR003347 JmjC_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR002138 Pept_C14_p10
IPR035892 C2_domain_sf
IPR000008 C2_dom
IPR030431 FAM189
IPR016024 ARM-type_fold
IPR017956 AT_hook_DNA-bd_motif
IPR039776 Pds5
IPR003349 JmjN
IPR001606 ARID_dom
IPR004198 Znf_C5HC2
IPR036431 ARID_dom_sf
IPR003347 JmjC_dom
Gene 3D
ProteinModelPortal
D7F175
D7F167
D7F162
D7F161
D7F163
D7F172
+ More
D7F168 D5LB39 A0A402EB12 A0A402EKC4 A0A2W1BNH4 A0A402EKB1 A0A402EST4 A0A402E768 A0A402EWU2 A0A402E7R7 A0A402EBP1 A0A402F9A7 A0A402ET04 A0A402EWP9 A0A402ECS2 A0A402ET28 A0A402F4E6 A0A402F9C1 A0A402F406 A0A402E7Q0 A0A402F125 A0A402EBC2 A0A402ELM7 A0A402FC20 A0A402FAG4 A0A402F575 A0A402EEU1 A0A402EWN2 A0A402F3F3 A0A402EKM3 A0A402EZW9 A0A402EVG7 A0A402EKK1 A0A402EWI9 A0A402EBI2 A0A402EXG4 A0A443QYR6 A0A402F382 A0A402EWK9 A0A402F9T4 A0A2H6KKM9 A0A402EWI8 A0A402ET45 A0A402EL35 A0A3S3P648 A0A3Q1LQN1 A0A402F7I8 A0A402F3Q6 H3B2Y3 A0A3Q1MLZ9 A0A3Q1M6J7 A0A402ER19 A0A3Q1MJK8 A0A0J7KGF7 A0A3Q1MI63 A0A3Q1MQ02 A0A3Q1M1T7 H2YWZ5 A0A3Q1MMW6 A0A3Q1N3Y9 A0A3Q1MGH6 A0A3Q1LWU3 A0A3Q1MP18 A0A3Q1M8D1 A0A3Q1LR18 A0A3Q1LUA6 A0A3Q1MA23 A0A3Q1LXQ5 A0A3Q1M527 A0A3Q1MDY8 A0A3Q1NE48 A0A3Q1MXT5 A0A3Q1MLI2 A0A402F6U7 A0A3Q1LI58 A0A3Q1N1P1 A0A3Q1N2X2 A0A3Q1MTQ8 A0A3Q1LNZ2 A0A3Q1MZJ0 A0A3Q1LX64 A0A3Q1MWB3 A0A3Q1LXF5 A0A3Q1MLV4 A0A3Q1ML42 A0A3Q1MNB9 A0A3Q1MM54 A0A3Q1LRD2 A0A3Q1LYT3 A0A3Q1LR52
D7F168 D5LB39 A0A402EB12 A0A402EKC4 A0A2W1BNH4 A0A402EKB1 A0A402EST4 A0A402E768 A0A402EWU2 A0A402E7R7 A0A402EBP1 A0A402F9A7 A0A402ET04 A0A402EWP9 A0A402ECS2 A0A402ET28 A0A402F4E6 A0A402F9C1 A0A402F406 A0A402E7Q0 A0A402F125 A0A402EBC2 A0A402ELM7 A0A402FC20 A0A402FAG4 A0A402F575 A0A402EEU1 A0A402EWN2 A0A402F3F3 A0A402EKM3 A0A402EZW9 A0A402EVG7 A0A402EKK1 A0A402EWI9 A0A402EBI2 A0A402EXG4 A0A443QYR6 A0A402F382 A0A402EWK9 A0A402F9T4 A0A2H6KKM9 A0A402EWI8 A0A402ET45 A0A402EL35 A0A3S3P648 A0A3Q1LQN1 A0A402F7I8 A0A402F3Q6 H3B2Y3 A0A3Q1MLZ9 A0A3Q1M6J7 A0A402ER19 A0A3Q1MJK8 A0A0J7KGF7 A0A3Q1MI63 A0A3Q1MQ02 A0A3Q1M1T7 H2YWZ5 A0A3Q1MMW6 A0A3Q1N3Y9 A0A3Q1MGH6 A0A3Q1LWU3 A0A3Q1MP18 A0A3Q1M8D1 A0A3Q1LR18 A0A3Q1LUA6 A0A3Q1MA23 A0A3Q1LXQ5 A0A3Q1M527 A0A3Q1MDY8 A0A3Q1NE48 A0A3Q1MXT5 A0A3Q1MLI2 A0A402F6U7 A0A3Q1LI58 A0A3Q1N1P1 A0A3Q1N2X2 A0A3Q1MTQ8 A0A3Q1LNZ2 A0A3Q1MZJ0 A0A3Q1LX64 A0A3Q1MWB3 A0A3Q1LXF5 A0A3Q1MLV4 A0A3Q1ML42 A0A3Q1MNB9 A0A3Q1MM54 A0A3Q1LRD2 A0A3Q1LYT3 A0A3Q1LR52
Ontologies
Topology
Subcellular location
Nucleus
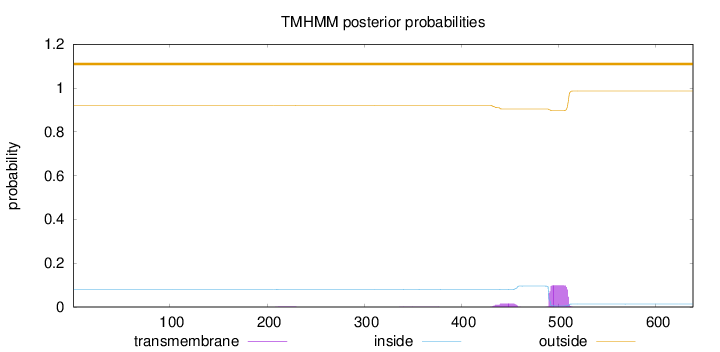
Length:
638
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.33383
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08105
outside
1 - 638
Population Genetic Test Statistics
Pi
3.175698
Theta
37.530812
Tajima's D
1.097043
CLR
0
CSRT
0.691165441727914
Interpretation
Uncertain
