Gene
KWMTBOMO08916
Pre Gene Modal
BGIBMGA007717
Annotation
PREDICTED:_zinc_finger_CCCH-type_with_G_patch_domain-containing_protein_[Amyelois_transitella]
Full name
Zinc finger CCCH-type with G patch domain-containing protein
Location in the cell
Nuclear Reliability : 4.214
Sequence
CDS
ATGGAAGACCTTGAATCGTCTTTAAATCAATACAAAGAACAAATTAAAATTGTGCAGCAATCCCTAGAAGCCACACATGACCTCAAAGAGAAAGAGTCTCTTTTAACACTACAATCTGATTTAAATCAACTAATTGAATTAACTCGAGAGAGCTTAGCAACAGCGACCAGCAAACATTTAAAATCCGATCTACAGAACAATGAGCAGAATAATTTGGATGAGGAATATGCTAGGTTCATGGCAGAAATGAATGACTCAAATGCATACAATGATGATAGCAAAAACAAAAATGATTCAGAAGAAAATAATGGCAACAGTGACATAGAGGATGACCTCTCTAGTTTATTGGGTATGAGGTGTGCTGTTTATCATACACACAAATGGGGTGGCCAGCCTAGTCTACACAATGCTATGGTCAGCTCGATACTGCCTAGGGCAGATGATGACCAATTCAATGATCTTAAGGTTCAAATATTATTTACTCATCCAACCCATGCAGAAATGTTGCCTTGTCCATTTTACCTAGATGGAGATTGTAAGTTTAGTGATGAACAATGCAGATATTCGCATGGTAATGTTGTACAACTGTCTGACCTAAAAGAAGCAATAGAACCGAATTTCAGTTCACTTAAATCCGGTAGCAGTATTTTGTTGAAATTGAAACCGCCAGATGATGAGAATGTAAGCATAACAAAAAAGTCTACAGAAAAATATCATCTATGGCATAGGGGAATAGTGAAATCTATAGACAATGAAAACCAATTGTGCACAGTTAAATTGGAGCACGGCGTTAAAACCGGCGAAAAAAGGAAGATGGGATCAGATGAAATTAATGTGCGCATCGAAGATATTTTCCCTCTTACCACCGTATGCGAAGATGATTCGGATTCCAGTGAAGATCTGAGCGACTCTGAATATTCCCATCAGAAAACAACACGAACGGAAATCGACAGACACGCGCTTATCGTCGAAAAGAGCTTACAGAATAATGCACCAGCTATGGGAGAGTGGGAGCGACATACTAGAGGTATAGGCTCGAAGATTATGTTCGCCATGGGCTATGTTGCGGGCTCCGGTTTGGGCGCCGCGGGCGAGGGGCGGACGCAGCCGGTTGAAGCGCGGGTGTTGCCCGTCGGGAAGAGCTTAGACCATTGCATGGCGATCAGTGAGAAACACGCCGGTCAGGATCCATTGAAGGTAACATTAACGGTTCTGATATAG
Protein
MEDLESSLNQYKEQIKIVQQSLEATHDLKEKESLLTLQSDLNQLIELTRESLATATSKHLKSDLQNNEQNNLDEEYARFMAEMNDSNAYNDDSKNKNDSEENNGNSDIEDDLSSLLGMRCAVYHTHKWGGQPSLHNAMVSSILPRADDDQFNDLKVQILFTHPTHAEMLPCPFYLDGDCKFSDEQCRYSHGNVVQLSDLKEAIEPNFSSLKSGSSILLKLKPPDDENVSITKKSTEKYHLWHRGIVKSIDNENQLCTVKLEHGVKTGEKRKMGSDEINVRIEDIFPLTTVCEDDSDSSEDLSDSEYSHQKTTRTEIDRHALIVEKSLQNNAPAMGEWERHTRGIGSKIMFAMGYVAGSGLGAAGEGRTQPVEARVLPVGKSLDHCMAISEKHAGQDPLKVTLTVLI
Summary
Description
Transcription repressor.
Keywords
Complete proteome
DNA-binding
Metal-binding
Nucleus
Reference proteome
Repressor
Transcription
Transcription regulation
Zinc
Zinc-finger
Feature
chain Zinc finger CCCH-type with G patch domain-containing protein
Uniprot
H9JDX0
A0A2H1WAK1
A0A2A4J0N8
A0A212EWW6
A0A194PIM3
A0A0L7KSW1
+ More
A0A1E1WS37 A0A0N1IJG9 A0A3S2LND6 A0A084W9U9 A0A310SRR2 A0A067QUN9 A0A2A3EGG4 A0A088AK31 A0A182LQX7 A0A182N6A0 A0A182KAE3 A0A182XGH3 E2BTC9 Q17CQ8 A0A154PI93 A0A182VLD2 A0A232F8Y2 A0A182PNC1 A0A1Y1MMT3 Q7PYU6 A0A1W4X748 A0A182Q6B9 A0A182JM81 A0A023EUK9 A0A182H3T6 A0A182GFG8 A0A2J7PIZ6 A0A182HZV3 E9IWB0 A0A2J7PIZ9 A0A0J7KDV6 W5JQ79 E2ARD4 A0A1B6LBU8 A0A182VRE0 A0A182SA81 A0A151I503 A0A0K8TRQ5 E0VVN3 A0A1B6DI03 A0A158NQC3 A0A182F7Y7 A0A182M3F4 A0A182RT79 A0A182XW29 F4WBB8 A0A151JPY2 A0A151JU37 A0A0L7R5F0 W8B6I9 A0A3L8DNJ6 B3MPC0 A0A0A1X518 B4KH32 A0A151WGF4 B4JCG4 A0A182U2V9 A0A1B6F6I4 A0A026WRJ3 Q29NF3 A0A3B0JX36 B4G7U3 B4M9F7 A0A131YTE7 A0A131XE51 T1HVF0 A0A131YRT5 X2JA03 Q9VL59 A0A224Z479 B3N8L3 A0A1E1XMK9 B4NYQ2 B4HWD7 A0A0P4VWB6 R7UKD3 L7M923 T1DSU9 A0A151IDY5 A0A0L0CGS9 A0A1I8M0Y0 A0A0J9QZN4 B4Q8A7 A0A1E1XB38 A0A1W4UUA2 T1IUR7 A0A2R5LDK1 A0A147BRI6 B7P636 A0A2S2R8N7 A0A146MDD8 A0A210Q1P6 A0A0Q9WWP5
A0A1E1WS37 A0A0N1IJG9 A0A3S2LND6 A0A084W9U9 A0A310SRR2 A0A067QUN9 A0A2A3EGG4 A0A088AK31 A0A182LQX7 A0A182N6A0 A0A182KAE3 A0A182XGH3 E2BTC9 Q17CQ8 A0A154PI93 A0A182VLD2 A0A232F8Y2 A0A182PNC1 A0A1Y1MMT3 Q7PYU6 A0A1W4X748 A0A182Q6B9 A0A182JM81 A0A023EUK9 A0A182H3T6 A0A182GFG8 A0A2J7PIZ6 A0A182HZV3 E9IWB0 A0A2J7PIZ9 A0A0J7KDV6 W5JQ79 E2ARD4 A0A1B6LBU8 A0A182VRE0 A0A182SA81 A0A151I503 A0A0K8TRQ5 E0VVN3 A0A1B6DI03 A0A158NQC3 A0A182F7Y7 A0A182M3F4 A0A182RT79 A0A182XW29 F4WBB8 A0A151JPY2 A0A151JU37 A0A0L7R5F0 W8B6I9 A0A3L8DNJ6 B3MPC0 A0A0A1X518 B4KH32 A0A151WGF4 B4JCG4 A0A182U2V9 A0A1B6F6I4 A0A026WRJ3 Q29NF3 A0A3B0JX36 B4G7U3 B4M9F7 A0A131YTE7 A0A131XE51 T1HVF0 A0A131YRT5 X2JA03 Q9VL59 A0A224Z479 B3N8L3 A0A1E1XMK9 B4NYQ2 B4HWD7 A0A0P4VWB6 R7UKD3 L7M923 T1DSU9 A0A151IDY5 A0A0L0CGS9 A0A1I8M0Y0 A0A0J9QZN4 B4Q8A7 A0A1E1XB38 A0A1W4UUA2 T1IUR7 A0A2R5LDK1 A0A147BRI6 B7P636 A0A2S2R8N7 A0A146MDD8 A0A210Q1P6 A0A0Q9WWP5
Pubmed
19121390
22118469
26354079
26227816
24438588
24845553
+ More
20966253 20798317 17510324 28648823 28004739 12364791 24945155 26483478 21282665 20920257 23761445 26369729 20566863 21347285 25244985 21719571 24495485 30249741 17994087 25830018 24508170 15632085 26830274 28049606 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 28797301 29209593 27129103 23254933 25576852 26108605 25315136 22936249 28503490 29652888 26823975 28812685
20966253 20798317 17510324 28648823 28004739 12364791 24945155 26483478 21282665 20920257 23761445 26369729 20566863 21347285 25244985 21719571 24495485 30249741 17994087 25830018 24508170 15632085 26830274 28049606 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 28797301 29209593 27129103 23254933 25576852 26108605 25315136 22936249 28503490 29652888 26823975 28812685
EMBL
BABH01002655
BABH01002656
ODYU01007381
SOQ50088.1
NWSH01004140
PCG65439.1
+ More
AGBW02011886 OWR45973.1 KQ459602 KPI93271.1 JTDY01006025 KOB66357.1 GDQN01001215 JAT89839.1 KQ459937 KPJ19227.1 RSAL01000049 RVE50404.1 ATLV01021917 KE525325 KFB46993.1 KQ759867 OAD62467.1 KK852930 KDR13643.1 KZ288256 PBC30574.1 GL450389 EFN81050.1 CH477305 KQ434923 KZC11527.1 NNAY01000700 OXU26919.1 GEZM01029655 JAV86010.1 AAAB01008987 AXCN02000277 GAPW01000977 JAC12621.1 JXUM01024830 KQ560702 KXJ81225.1 JXUM01059565 KQ562060 KXJ76781.1 NEVH01024952 PNF16311.1 APCN01002014 GL766483 EFZ15142.1 PNF16317.1 LBMM01009110 KMQ88384.1 ADMH02000423 ETN66567.1 GL442048 EFN64005.1 GEBQ01018815 JAT21162.1 KQ976451 KYM85601.1 GDAI01000780 JAI16823.1 DS235812 EEB17439.1 GEDC01011994 JAS25304.1 ADTU01023123 ADTU01023124 ADTU01023125 AXCM01000396 GL888062 EGI68474.1 KQ978756 KYN28575.1 KQ981788 KYN35793.1 KQ414654 KOC65986.1 GAMC01012338 JAB94217.1 QOIP01000006 RLU22015.1 CH902620 GBXI01007873 JAD06419.1 CH933807 KQ983176 KYQ46904.1 CH916368 GECZ01024306 JAS45463.1 KK107128 EZA58311.1 CH379060 OUUW01000010 SPP85633.1 CH479180 CH940654 GEDV01007376 JAP81181.1 GEFH01004223 JAP64358.1 ACPB03015772 GEDV01007377 JAP81180.1 AE014134 AHN54302.1 AY058650 GFPF01013471 MAA24617.1 CH954177 GFAA01003108 JAU00327.1 CM000157 CH480818 GDKW01000190 JAI56405.1 AMQN01001209 KB300511 ELU06660.1 GACK01005450 JAA59584.1 GAMD01001319 JAB00272.1 KQ977901 KYM98891.1 JRES01000418 KNC31441.1 CM002910 KMY89373.1 CM000361 GFAC01002728 JAT96460.1 JH431553 GGLE01003379 MBY07505.1 GEGO01002038 JAR93366.1 ABJB010334862 ABJB010663500 DS644093 EEC02058.1 GGMS01017224 MBY86427.1 GDHC01001873 JAQ16756.1 NEDP02005240 OWF42647.1 CH969605 KRG00459.1
AGBW02011886 OWR45973.1 KQ459602 KPI93271.1 JTDY01006025 KOB66357.1 GDQN01001215 JAT89839.1 KQ459937 KPJ19227.1 RSAL01000049 RVE50404.1 ATLV01021917 KE525325 KFB46993.1 KQ759867 OAD62467.1 KK852930 KDR13643.1 KZ288256 PBC30574.1 GL450389 EFN81050.1 CH477305 KQ434923 KZC11527.1 NNAY01000700 OXU26919.1 GEZM01029655 JAV86010.1 AAAB01008987 AXCN02000277 GAPW01000977 JAC12621.1 JXUM01024830 KQ560702 KXJ81225.1 JXUM01059565 KQ562060 KXJ76781.1 NEVH01024952 PNF16311.1 APCN01002014 GL766483 EFZ15142.1 PNF16317.1 LBMM01009110 KMQ88384.1 ADMH02000423 ETN66567.1 GL442048 EFN64005.1 GEBQ01018815 JAT21162.1 KQ976451 KYM85601.1 GDAI01000780 JAI16823.1 DS235812 EEB17439.1 GEDC01011994 JAS25304.1 ADTU01023123 ADTU01023124 ADTU01023125 AXCM01000396 GL888062 EGI68474.1 KQ978756 KYN28575.1 KQ981788 KYN35793.1 KQ414654 KOC65986.1 GAMC01012338 JAB94217.1 QOIP01000006 RLU22015.1 CH902620 GBXI01007873 JAD06419.1 CH933807 KQ983176 KYQ46904.1 CH916368 GECZ01024306 JAS45463.1 KK107128 EZA58311.1 CH379060 OUUW01000010 SPP85633.1 CH479180 CH940654 GEDV01007376 JAP81181.1 GEFH01004223 JAP64358.1 ACPB03015772 GEDV01007377 JAP81180.1 AE014134 AHN54302.1 AY058650 GFPF01013471 MAA24617.1 CH954177 GFAA01003108 JAU00327.1 CM000157 CH480818 GDKW01000190 JAI56405.1 AMQN01001209 KB300511 ELU06660.1 GACK01005450 JAA59584.1 GAMD01001319 JAB00272.1 KQ977901 KYM98891.1 JRES01000418 KNC31441.1 CM002910 KMY89373.1 CM000361 GFAC01002728 JAT96460.1 JH431553 GGLE01003379 MBY07505.1 GEGO01002038 JAR93366.1 ABJB010334862 ABJB010663500 DS644093 EEC02058.1 GGMS01017224 MBY86427.1 GDHC01001873 JAQ16756.1 NEDP02005240 OWF42647.1 CH969605 KRG00459.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037510
UP000053240
+ More
UP000283053 UP000030765 UP000027135 UP000242457 UP000005203 UP000075882 UP000075884 UP000075881 UP000076407 UP000008237 UP000008820 UP000076502 UP000075903 UP000215335 UP000075885 UP000007062 UP000192223 UP000075886 UP000075880 UP000069940 UP000249989 UP000235965 UP000075840 UP000036403 UP000000673 UP000000311 UP000075920 UP000075901 UP000078540 UP000009046 UP000005205 UP000069272 UP000075883 UP000075900 UP000076408 UP000007755 UP000078492 UP000078541 UP000053825 UP000279307 UP000007801 UP000009192 UP000075809 UP000001070 UP000075902 UP000053097 UP000001819 UP000268350 UP000008744 UP000008792 UP000015103 UP000000803 UP000008711 UP000002282 UP000001292 UP000014760 UP000078542 UP000037069 UP000095301 UP000000304 UP000192221 UP000001555 UP000242188 UP000007798
UP000283053 UP000030765 UP000027135 UP000242457 UP000005203 UP000075882 UP000075884 UP000075881 UP000076407 UP000008237 UP000008820 UP000076502 UP000075903 UP000215335 UP000075885 UP000007062 UP000192223 UP000075886 UP000075880 UP000069940 UP000249989 UP000235965 UP000075840 UP000036403 UP000000673 UP000000311 UP000075920 UP000075901 UP000078540 UP000009046 UP000005205 UP000069272 UP000075883 UP000075900 UP000076408 UP000007755 UP000078492 UP000078541 UP000053825 UP000279307 UP000007801 UP000009192 UP000075809 UP000001070 UP000075902 UP000053097 UP000001819 UP000268350 UP000008744 UP000008792 UP000015103 UP000000803 UP000008711 UP000002282 UP000001292 UP000014760 UP000078542 UP000037069 UP000095301 UP000000304 UP000192221 UP000001555 UP000242188 UP000007798
ProteinModelPortal
H9JDX0
A0A2H1WAK1
A0A2A4J0N8
A0A212EWW6
A0A194PIM3
A0A0L7KSW1
+ More
A0A1E1WS37 A0A0N1IJG9 A0A3S2LND6 A0A084W9U9 A0A310SRR2 A0A067QUN9 A0A2A3EGG4 A0A088AK31 A0A182LQX7 A0A182N6A0 A0A182KAE3 A0A182XGH3 E2BTC9 Q17CQ8 A0A154PI93 A0A182VLD2 A0A232F8Y2 A0A182PNC1 A0A1Y1MMT3 Q7PYU6 A0A1W4X748 A0A182Q6B9 A0A182JM81 A0A023EUK9 A0A182H3T6 A0A182GFG8 A0A2J7PIZ6 A0A182HZV3 E9IWB0 A0A2J7PIZ9 A0A0J7KDV6 W5JQ79 E2ARD4 A0A1B6LBU8 A0A182VRE0 A0A182SA81 A0A151I503 A0A0K8TRQ5 E0VVN3 A0A1B6DI03 A0A158NQC3 A0A182F7Y7 A0A182M3F4 A0A182RT79 A0A182XW29 F4WBB8 A0A151JPY2 A0A151JU37 A0A0L7R5F0 W8B6I9 A0A3L8DNJ6 B3MPC0 A0A0A1X518 B4KH32 A0A151WGF4 B4JCG4 A0A182U2V9 A0A1B6F6I4 A0A026WRJ3 Q29NF3 A0A3B0JX36 B4G7U3 B4M9F7 A0A131YTE7 A0A131XE51 T1HVF0 A0A131YRT5 X2JA03 Q9VL59 A0A224Z479 B3N8L3 A0A1E1XMK9 B4NYQ2 B4HWD7 A0A0P4VWB6 R7UKD3 L7M923 T1DSU9 A0A151IDY5 A0A0L0CGS9 A0A1I8M0Y0 A0A0J9QZN4 B4Q8A7 A0A1E1XB38 A0A1W4UUA2 T1IUR7 A0A2R5LDK1 A0A147BRI6 B7P636 A0A2S2R8N7 A0A146MDD8 A0A210Q1P6 A0A0Q9WWP5
A0A1E1WS37 A0A0N1IJG9 A0A3S2LND6 A0A084W9U9 A0A310SRR2 A0A067QUN9 A0A2A3EGG4 A0A088AK31 A0A182LQX7 A0A182N6A0 A0A182KAE3 A0A182XGH3 E2BTC9 Q17CQ8 A0A154PI93 A0A182VLD2 A0A232F8Y2 A0A182PNC1 A0A1Y1MMT3 Q7PYU6 A0A1W4X748 A0A182Q6B9 A0A182JM81 A0A023EUK9 A0A182H3T6 A0A182GFG8 A0A2J7PIZ6 A0A182HZV3 E9IWB0 A0A2J7PIZ9 A0A0J7KDV6 W5JQ79 E2ARD4 A0A1B6LBU8 A0A182VRE0 A0A182SA81 A0A151I503 A0A0K8TRQ5 E0VVN3 A0A1B6DI03 A0A158NQC3 A0A182F7Y7 A0A182M3F4 A0A182RT79 A0A182XW29 F4WBB8 A0A151JPY2 A0A151JU37 A0A0L7R5F0 W8B6I9 A0A3L8DNJ6 B3MPC0 A0A0A1X518 B4KH32 A0A151WGF4 B4JCG4 A0A182U2V9 A0A1B6F6I4 A0A026WRJ3 Q29NF3 A0A3B0JX36 B4G7U3 B4M9F7 A0A131YTE7 A0A131XE51 T1HVF0 A0A131YRT5 X2JA03 Q9VL59 A0A224Z479 B3N8L3 A0A1E1XMK9 B4NYQ2 B4HWD7 A0A0P4VWB6 R7UKD3 L7M923 T1DSU9 A0A151IDY5 A0A0L0CGS9 A0A1I8M0Y0 A0A0J9QZN4 B4Q8A7 A0A1E1XB38 A0A1W4UUA2 T1IUR7 A0A2R5LDK1 A0A147BRI6 B7P636 A0A2S2R8N7 A0A146MDD8 A0A210Q1P6 A0A0Q9WWP5
PDB
4II1
E-value=3.13941e-11,
Score=165
Ontologies
KEGG
GO
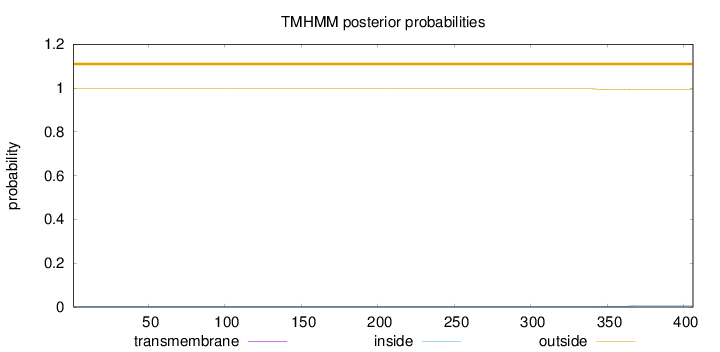
Topology
Subcellular location
Nucleus
Length:
406
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08546
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00368
outside
1 - 406
Population Genetic Test Statistics
Pi
223.897796
Theta
158.532568
Tajima's D
1.005443
CLR
0.23632
CSRT
0.654267286635668
Interpretation
Uncertain
