Gene
KWMTBOMO08914
Annotation
integrase_core_domain_protein_[Lasius_niger]
Location in the cell
Nuclear Reliability : 3.343
Sequence
CDS
ATGCATTATAGCGGTACTCCTGCTGAGAAACGCGAAGGTCTTACTCCAGAACCTGAAACAGAACAAATTGAAATTCCAGAAAAAAGAAACAACATTAATGAGAACTTACCTTTTCAGGAACCTAACCCCCTTTATTCCAGCTCTAGTCAAGATCATGTTGATGAACCATCTCCCTCAAGTGGAAGACCACAAAGGGACGTGAGACCACCAGCATGGACCAAAGATTATGAGTTAAGTCGCTGCGTGACCATGTTTACGTCAGAAAATAATGACGACTATGACTGCTCTTTACTAACATACAAAGAAGCTGTGACTGGCACCGACAGACGAAATTGGGAGAAGGCGATACAATCTGAAATGGAATCATTACAGAAAAATAACACCTGGGTATTAGTAGACAAGTCAGAAGCCAAAGGACACAAAATACTGTCGAACAAATGGGTATTTCGGATTAAAGATGACGGTACGTACAAGGCTAGATTAGTCGTACGTGGATGCGAGCAGAAAGGAAACCTGGATTTCGAAGACATTTATAGCCCAGTAGTAAGTCAATCTGCATTAAAAACTTTATTAGCAATTGCCGCATCTAAATCTTACTACTTCATGACATTTGACATCAAGACAGCTTTCTTATACGGAGAGTTATCAGATGAAGTTTATATGAAATTGCCTGTTGGCTATGAGACTAACTGCAATACTACTGAAGTAGTTTGCCGATTAAAAAAATCTTTGTACGGATTAAAACAAGCTCCATTTACATGGAACAAAACTTTTACCCAAGCGATGTTTGATTTGGGATTCAAGACAATCAAAACAGATCGATGCGTTTTCATAAACGAAGATAAATCAATAATAATCGGACTATTCGTTGATGATGGACTTGTCATAGGAAAAGATAAGAAAAAGATCGAAGCTATTTTGAAGAAACTTGAAACCAAATTTCAAATGAAAAAGAATGAAAATCCAAGTACTTATCTGGGTATGGAAATAAAAAACTCAAAAGAAGGAATCATTCTTACTCAGAGATCCTACATAAATGACGTACTGGAAAAATACAACATGAAAAATGCCAAACCTTGTGACACGCCCGCAAGTAGCGATTCAAAGTCGATACAGGAAACTGCCCAGGAGAAGGAATATCCCTATAGGGAAGCAGTGGGAAGTTTACTGTACCTGTCTACCAGAACCAGACCAGATATCGCACAAGCGGTCAATCTAGTTAGCAGGAAGGTCGAAAACCCCACTAGAGAAGACGTTACAAAATTGAAGAAAATATTCAAATATTTAGTGGGAACCAAACATAAAGGCATTTTATTCAAACGAAACAAACCGATAGAGAGCATCGATGCTTACAGCGACTCGGATTATGCTGGAGACCCATTGACGCGAAGAAGCACTTCTGGTTCCGTGCTAATGATAGCCGATGGACCAATATCATGGGCATCCAAGAGGCAGCCTATAGTAGCGCTATCGTCGACCGAAGCCGAGTACATAGCCGCAGCCGACTGTTGCAAAGAGGCATTGTATCTCAAATCTCTATTACAGGAAATAACCGGTATGGACATAAAAATAAATCTACATGTTGATAACCAAAGTTCGATTCAACTTGTGAAGTCTGGATCCTTCAACAAAAGGTCTAAGCACATAGACGTGCGGTACCATTTTATACATGAACACTTCGTGCATGGATTAATAAATATTAGCTACTGTCCCACAGAAATTCAGTTAGCTGACGTAATGACAAAACCATTGTCAAAAGTAAAATTTGAAAGACACTGTGCAAGATTAATGCATGTATAG
Protein
MHYSGTPAEKREGLTPEPETEQIEIPEKRNNINENLPFQEPNPLYSSSSQDHVDEPSPSSGRPQRDVRPPAWTKDYELSRCVTMFTSENNDDYDCSLLTYKEAVTGTDRRNWEKAIQSEMESLQKNNTWVLVDKSEAKGHKILSNKWVFRIKDDGTYKARLVVRGCEQKGNLDFEDIYSPVVSQSALKTLLAIAASKSYYFMTFDIKTAFLYGELSDEVYMKLPVGYETNCNTTEVVCRLKKSLYGLKQAPFTWNKTFTQAMFDLGFKTIKTDRCVFINEDKSIIIGLFVDDGLVIGKDKKKIEAILKKLETKFQMKKNENPSTYLGMEIKNSKEGIILTQRSYINDVLEKYNMKNAKPCDTPASSDSKSIQETAQEKEYPYREAVGSLLYLSTRTRPDIAQAVNLVSRKVENPTREDVTKLKKIFKYLVGTKHKGILFKRNKPIESIDAYSDSDYAGDPLTRRSTSGSVLMIADGPISWASKRQPIVALSSTEAEYIAAADCCKEALYLKSLLQEITGMDIKINLHVDNQSSIQLVKSGSFNKRSKHIDVRYHFIHEHFVHGLINISYCPTEIQLADVMTKPLSKVKFERHCARLMHV
Summary
Uniprot
A0A0A9YCF4
A0A0V0G477
A0A1B6C2D3
A0A0A9WDS7
A0A0A9W6J9
A0A0A9Z2D6
+ More
A0A0A9WIY3 A0A0A9YZ93 A0A0A9X817 A0A146M489 A0A0A9XEU3 A0A0A9YMV4 A0A0A9X3Z4 A0A1B6FNY3 A0A1Y1LVN7 A0A2S2NXV8 A0A146LHR9 X1X8M7 A0A438BTG2 X1XTZ0 A0A0K8TKJ8 X1WLH4 A0A139WJX6 A0A2N9GRB2 A0A0V1NCR1 A0A0J7KE66 A0A1Y1KNM9 A0A1Y1N3G5 A0A2N9HJ76 A0A085MTW9 A0A1Y1NM77 A0A438E2Y0 A0A2N9ELY1 A0A438I3N7 B8YLY7 A0A438E432 A0A1Y1MYL8 A0A2N9FQJ1 A0A2N9FV67 A0A1Y1MYL6 A0A2N9FRN5 A0A2N9GCE7 A0A2N9I8Q5 A0A2N9EK90 A0A2N9EIR6 A0A2N9I6N4 A0A2N9ID98 A0A2N9GA80 A0A2N9FZ01 A0A2N9FW82 A0A2N9H6B2 A0A151R6U8 A0A2N9GGS7 A0A0V0WRK3 A0A151R2G6 A0A151SDY6 A0A2N9ETC8 A0A2N9I7V5 A0A2N9HKJ4 A0A0J7K725 A0A151TKT1 A0A2N9IGN4 A0A151RJN4 A0A2N9H0T6 A0A2N9GRZ0 B8YLY3 A0A151TNJ8 A0A2N9GBF3 A0A2N9GBS9 B8YLY6 A0A0V1HMN9 A0A2N9IUP8 B8YLY5 A0A182H8F8 A0A2N9EE21 A0A2N9H2E5 A0A0V1IW77 A0A2N9H4V1 A0A2N9I1P0 B8YLY4 A0A1Y1MYP5 A0A0J7KRG1 A0A0V0YKY1 A0A151UC72 A0A151UBW1 A0A1J3DEI6 A0A151T7T6 A0A2N9I9F7 A0A1Y1M571 A0A438HJE1 A0A3S3PPW2 A0A0V1M1K0 H6U1F0 H9JF18 H6U1E9 A0A0V1B5K0 A0A0V1B5A2 A0A2N9F2B1
A0A0A9WIY3 A0A0A9YZ93 A0A0A9X817 A0A146M489 A0A0A9XEU3 A0A0A9YMV4 A0A0A9X3Z4 A0A1B6FNY3 A0A1Y1LVN7 A0A2S2NXV8 A0A146LHR9 X1X8M7 A0A438BTG2 X1XTZ0 A0A0K8TKJ8 X1WLH4 A0A139WJX6 A0A2N9GRB2 A0A0V1NCR1 A0A0J7KE66 A0A1Y1KNM9 A0A1Y1N3G5 A0A2N9HJ76 A0A085MTW9 A0A1Y1NM77 A0A438E2Y0 A0A2N9ELY1 A0A438I3N7 B8YLY7 A0A438E432 A0A1Y1MYL8 A0A2N9FQJ1 A0A2N9FV67 A0A1Y1MYL6 A0A2N9FRN5 A0A2N9GCE7 A0A2N9I8Q5 A0A2N9EK90 A0A2N9EIR6 A0A2N9I6N4 A0A2N9ID98 A0A2N9GA80 A0A2N9FZ01 A0A2N9FW82 A0A2N9H6B2 A0A151R6U8 A0A2N9GGS7 A0A0V0WRK3 A0A151R2G6 A0A151SDY6 A0A2N9ETC8 A0A2N9I7V5 A0A2N9HKJ4 A0A0J7K725 A0A151TKT1 A0A2N9IGN4 A0A151RJN4 A0A2N9H0T6 A0A2N9GRZ0 B8YLY3 A0A151TNJ8 A0A2N9GBF3 A0A2N9GBS9 B8YLY6 A0A0V1HMN9 A0A2N9IUP8 B8YLY5 A0A182H8F8 A0A2N9EE21 A0A2N9H2E5 A0A0V1IW77 A0A2N9H4V1 A0A2N9I1P0 B8YLY4 A0A1Y1MYP5 A0A0J7KRG1 A0A0V0YKY1 A0A151UC72 A0A151UBW1 A0A1J3DEI6 A0A151T7T6 A0A2N9I9F7 A0A1Y1M571 A0A438HJE1 A0A3S3PPW2 A0A0V1M1K0 H6U1F0 H9JF18 H6U1E9 A0A0V1B5K0 A0A0V1B5A2 A0A2N9F2B1
Pubmed
EMBL
GBHO01016409
JAG27195.1
GECL01003258
JAP02866.1
GEDC01029908
JAS07390.1
+ More
GBHO01040574 JAG03030.1 GBHO01040573 JAG03031.1 GBHO01006096 JAG37508.1 GBHO01035865 JAG07739.1 GBHO01035869 GBHO01006095 JAG07735.1 JAG37509.1 GBHO01028636 GBHO01028635 JAG14968.1 JAG14969.1 GDHC01005053 JAQ13576.1 GBHO01024362 JAG19242.1 GBHO01012744 JAG30860.1 GBHO01031779 JAG11825.1 GECZ01017843 JAS51926.1 GEZM01048830 JAV76370.1 GGMR01009345 MBY21964.1 GDHC01010945 JAQ07684.1 ABLF02042027 QGNW01002623 RVW14265.1 ABLF02043571 ABLF02045585 ABLF02057174 ABLF02063637 ABLF02063640 GDAI01002967 JAI14636.1 ABLF02026843 KQ971338 KYB28117.1 OIVN01002268 SPD02135.1 JYDM01000512 KRZ81798.1 LBMM01008978 KMQ88491.1 GEZM01078128 GEZM01078127 JAV63002.1 GEZM01013586 JAV92451.1 OIVN01003897 SPD14257.1 KL367655 KFD60665.1 GEZM01002084 JAV97376.1 QGNW01001416 RVW42052.1 OIVN01000183 SPC75833.1 QGNW01000147 RVW91307.1 FJ544857 ACL97387.1 QGNW01001404 RVW42505.1 GEZM01017328 JAV90763.1 OIVN01001052 SPC89229.1 OIVN01001196 SPC91020.1 GEZM01017329 JAV90762.1 OIVN01001091 SPC89740.1 OIVN01001729 SPC97049.1 OIVN01005024 SPD20514.1 OIVN01000147 SPC75222.1 OIVN01000116 SPC74628.1 OIVN01004879 SPD19723.1 OIVN01005312 SPD21871.1 OIVN01001666 SPC96375.1 OIVN01001309 SPC92453.1 OIVN01001520 SPC94877.1 OIVN01002904 SPD07378.1 KQ484038 KYP38095.1 OIVN01001890 SPC98658.1 JYDK01000067 KRX78390.1 KQ484186 KYP36645.1 KQ483417 KYP53032.1 OIVN01000557 SPC82046.1 OIVN01004910 SPD19931.1 OIVN01003569 SPD12194.1 LBMM01012536 KMQ86167.1 CM003607 KYP67664.1 OIVN01005657 SPD23465.1 KQ483702 KYP42748.1 OIVN01002680 SPD05612.1 OIVN01002256 SPD02034.1 FJ544851 ACL97383.1 CM003606 KYP68590.1 OIVN01001669 SPC96464.1 OIVN01001707 SPC96839.1 FJ544856 ACL97386.1 JYDS01000349 KRZ11769.1 OIVN01006259 SPD29117.1 FJ544854 ACL97385.1 JXUM01118474 KQ566190 KXJ70335.1 OIVN01000036 SPC73022.1 OIVN01002725 SPD05963.1 JYDS01000077 KRZ26987.1 OIVN01002854 SPD06945.1 OIVN01004603 SPD18338.1 FJ544853 ACL97384.1 GEZM01017327 JAV90764.1 LBMM01003976 KMQ92933.1 JYDU01000007 KRY00737.1 CM003603 KYP76811.1 KYP76810.1 GEVI01015824 JAU16496.1 CM003610 KYP63120.1 OIVN01005059 SPD20680.1 GEZM01040221 GEZM01040220 JAV80932.1 QGNW01000214 RVW84557.1 NCKU01004452 RWS05910.1 JYDO01000312 KRZ65659.1 JN402333 AFB73912.1 BABH01033959 BABH01033960 JN402332 AFB73911.1 JYDH01000102 KRY32233.1 KRY32232.1 OIVN01000758 SPC85117.1
GBHO01040574 JAG03030.1 GBHO01040573 JAG03031.1 GBHO01006096 JAG37508.1 GBHO01035865 JAG07739.1 GBHO01035869 GBHO01006095 JAG07735.1 JAG37509.1 GBHO01028636 GBHO01028635 JAG14968.1 JAG14969.1 GDHC01005053 JAQ13576.1 GBHO01024362 JAG19242.1 GBHO01012744 JAG30860.1 GBHO01031779 JAG11825.1 GECZ01017843 JAS51926.1 GEZM01048830 JAV76370.1 GGMR01009345 MBY21964.1 GDHC01010945 JAQ07684.1 ABLF02042027 QGNW01002623 RVW14265.1 ABLF02043571 ABLF02045585 ABLF02057174 ABLF02063637 ABLF02063640 GDAI01002967 JAI14636.1 ABLF02026843 KQ971338 KYB28117.1 OIVN01002268 SPD02135.1 JYDM01000512 KRZ81798.1 LBMM01008978 KMQ88491.1 GEZM01078128 GEZM01078127 JAV63002.1 GEZM01013586 JAV92451.1 OIVN01003897 SPD14257.1 KL367655 KFD60665.1 GEZM01002084 JAV97376.1 QGNW01001416 RVW42052.1 OIVN01000183 SPC75833.1 QGNW01000147 RVW91307.1 FJ544857 ACL97387.1 QGNW01001404 RVW42505.1 GEZM01017328 JAV90763.1 OIVN01001052 SPC89229.1 OIVN01001196 SPC91020.1 GEZM01017329 JAV90762.1 OIVN01001091 SPC89740.1 OIVN01001729 SPC97049.1 OIVN01005024 SPD20514.1 OIVN01000147 SPC75222.1 OIVN01000116 SPC74628.1 OIVN01004879 SPD19723.1 OIVN01005312 SPD21871.1 OIVN01001666 SPC96375.1 OIVN01001309 SPC92453.1 OIVN01001520 SPC94877.1 OIVN01002904 SPD07378.1 KQ484038 KYP38095.1 OIVN01001890 SPC98658.1 JYDK01000067 KRX78390.1 KQ484186 KYP36645.1 KQ483417 KYP53032.1 OIVN01000557 SPC82046.1 OIVN01004910 SPD19931.1 OIVN01003569 SPD12194.1 LBMM01012536 KMQ86167.1 CM003607 KYP67664.1 OIVN01005657 SPD23465.1 KQ483702 KYP42748.1 OIVN01002680 SPD05612.1 OIVN01002256 SPD02034.1 FJ544851 ACL97383.1 CM003606 KYP68590.1 OIVN01001669 SPC96464.1 OIVN01001707 SPC96839.1 FJ544856 ACL97386.1 JYDS01000349 KRZ11769.1 OIVN01006259 SPD29117.1 FJ544854 ACL97385.1 JXUM01118474 KQ566190 KXJ70335.1 OIVN01000036 SPC73022.1 OIVN01002725 SPD05963.1 JYDS01000077 KRZ26987.1 OIVN01002854 SPD06945.1 OIVN01004603 SPD18338.1 FJ544853 ACL97384.1 GEZM01017327 JAV90764.1 LBMM01003976 KMQ92933.1 JYDU01000007 KRY00737.1 CM003603 KYP76811.1 KYP76810.1 GEVI01015824 JAU16496.1 CM003610 KYP63120.1 OIVN01005059 SPD20680.1 GEZM01040221 GEZM01040220 JAV80932.1 QGNW01000214 RVW84557.1 NCKU01004452 RWS05910.1 JYDO01000312 KRZ65659.1 JN402333 AFB73912.1 BABH01033959 BABH01033960 JN402332 AFB73911.1 JYDH01000102 KRY32233.1 KRY32232.1 OIVN01000758 SPC85117.1
Proteomes
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A0A9YCF4
A0A0V0G477
A0A1B6C2D3
A0A0A9WDS7
A0A0A9W6J9
A0A0A9Z2D6
+ More
A0A0A9WIY3 A0A0A9YZ93 A0A0A9X817 A0A146M489 A0A0A9XEU3 A0A0A9YMV4 A0A0A9X3Z4 A0A1B6FNY3 A0A1Y1LVN7 A0A2S2NXV8 A0A146LHR9 X1X8M7 A0A438BTG2 X1XTZ0 A0A0K8TKJ8 X1WLH4 A0A139WJX6 A0A2N9GRB2 A0A0V1NCR1 A0A0J7KE66 A0A1Y1KNM9 A0A1Y1N3G5 A0A2N9HJ76 A0A085MTW9 A0A1Y1NM77 A0A438E2Y0 A0A2N9ELY1 A0A438I3N7 B8YLY7 A0A438E432 A0A1Y1MYL8 A0A2N9FQJ1 A0A2N9FV67 A0A1Y1MYL6 A0A2N9FRN5 A0A2N9GCE7 A0A2N9I8Q5 A0A2N9EK90 A0A2N9EIR6 A0A2N9I6N4 A0A2N9ID98 A0A2N9GA80 A0A2N9FZ01 A0A2N9FW82 A0A2N9H6B2 A0A151R6U8 A0A2N9GGS7 A0A0V0WRK3 A0A151R2G6 A0A151SDY6 A0A2N9ETC8 A0A2N9I7V5 A0A2N9HKJ4 A0A0J7K725 A0A151TKT1 A0A2N9IGN4 A0A151RJN4 A0A2N9H0T6 A0A2N9GRZ0 B8YLY3 A0A151TNJ8 A0A2N9GBF3 A0A2N9GBS9 B8YLY6 A0A0V1HMN9 A0A2N9IUP8 B8YLY5 A0A182H8F8 A0A2N9EE21 A0A2N9H2E5 A0A0V1IW77 A0A2N9H4V1 A0A2N9I1P0 B8YLY4 A0A1Y1MYP5 A0A0J7KRG1 A0A0V0YKY1 A0A151UC72 A0A151UBW1 A0A1J3DEI6 A0A151T7T6 A0A2N9I9F7 A0A1Y1M571 A0A438HJE1 A0A3S3PPW2 A0A0V1M1K0 H6U1F0 H9JF18 H6U1E9 A0A0V1B5K0 A0A0V1B5A2 A0A2N9F2B1
A0A0A9WIY3 A0A0A9YZ93 A0A0A9X817 A0A146M489 A0A0A9XEU3 A0A0A9YMV4 A0A0A9X3Z4 A0A1B6FNY3 A0A1Y1LVN7 A0A2S2NXV8 A0A146LHR9 X1X8M7 A0A438BTG2 X1XTZ0 A0A0K8TKJ8 X1WLH4 A0A139WJX6 A0A2N9GRB2 A0A0V1NCR1 A0A0J7KE66 A0A1Y1KNM9 A0A1Y1N3G5 A0A2N9HJ76 A0A085MTW9 A0A1Y1NM77 A0A438E2Y0 A0A2N9ELY1 A0A438I3N7 B8YLY7 A0A438E432 A0A1Y1MYL8 A0A2N9FQJ1 A0A2N9FV67 A0A1Y1MYL6 A0A2N9FRN5 A0A2N9GCE7 A0A2N9I8Q5 A0A2N9EK90 A0A2N9EIR6 A0A2N9I6N4 A0A2N9ID98 A0A2N9GA80 A0A2N9FZ01 A0A2N9FW82 A0A2N9H6B2 A0A151R6U8 A0A2N9GGS7 A0A0V0WRK3 A0A151R2G6 A0A151SDY6 A0A2N9ETC8 A0A2N9I7V5 A0A2N9HKJ4 A0A0J7K725 A0A151TKT1 A0A2N9IGN4 A0A151RJN4 A0A2N9H0T6 A0A2N9GRZ0 B8YLY3 A0A151TNJ8 A0A2N9GBF3 A0A2N9GBS9 B8YLY6 A0A0V1HMN9 A0A2N9IUP8 B8YLY5 A0A182H8F8 A0A2N9EE21 A0A2N9H2E5 A0A0V1IW77 A0A2N9H4V1 A0A2N9I1P0 B8YLY4 A0A1Y1MYP5 A0A0J7KRG1 A0A0V0YKY1 A0A151UC72 A0A151UBW1 A0A1J3DEI6 A0A151T7T6 A0A2N9I9F7 A0A1Y1M571 A0A438HJE1 A0A3S3PPW2 A0A0V1M1K0 H6U1F0 H9JF18 H6U1E9 A0A0V1B5K0 A0A0V1B5A2 A0A2N9F2B1
Ontologies
PANTHER
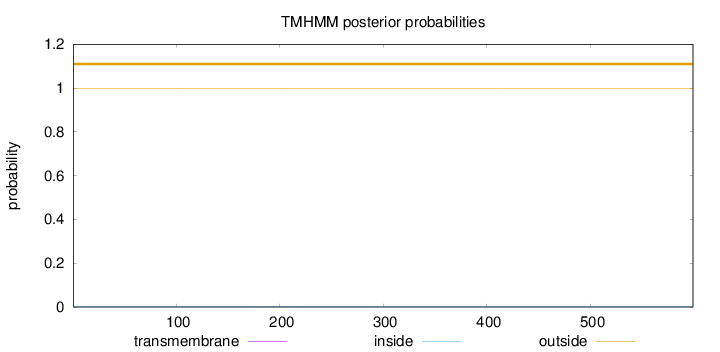
Topology
Length:
599
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01259
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00141
outside
1 - 599
Population Genetic Test Statistics
Pi
190.734246
Theta
125.172389
Tajima's D
0.177561
CLR
0
CSRT
0.417079146042698
Interpretation
Uncertain
