Gene
KWMTBOMO08913 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007817
Annotation
PREDICTED:_uncharacterized_protein_LOC100862770_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.824
Sequence
CDS
ATGACAATGTTTTCATGCATAACCTGTAAGGTGTTGTTTAACACATCCGAATTACAACGTGAACATTATAAATTAGATTGGCATAGATATAATTTAAAAAGAAAAGTAGCGTCTATTGAACCTGTCACCCTTGAGGAGTTTGAAGAGAGAGCTAAAGAACATAGAGAAAGTCAAAATGAAAAGCAAGATGATTCACAATACTGCCAATGCTGCTCAAAGTTATTCAGCACCAAGAACTCTTATAACAATCATTTGAATAGTAAGAAACACAAAGTGAGTGTTGAAAGATACACAGAAAGTCAAAAGGATCAAGAGAACAGCGGTCAATCTGATACAGACAGTTTTGTTAAAGTTGAATGTACTACTGGCTTAACCAACGAACGGAGTAAATTTGTCGTTGTGAACACTACTGATTCTGGAGAAGATATAGAAACTGACTCTGAAATAGAGGAGCTAGATTCCGACGAATGGGATGAGTGTCGCATCCAAGAAAGTGATTCACTTATCAAACCCCGGGATTGTTTGTTCTGTGTACATCACAGTAAGAATATGGTAAAAAACTTAAAGCACATGTCAGAAGTACATTCGTTCTTCATTCCTGATGTAGAATTTTGTATAAATATTAGAGGTCTGCTGTTGTACTTAGGAGAAAAGATATCACAGGGATATATGTGCCTATGGTGTAATGACACTGGAAGGACATTCTACTCAATGGAAGCAGCACGGGCCCACATGATTGATAAGGGACATTGCAAGATGTTACACGAAGGTCTTGCTTTAGCGGAGTATGCAGACTACTATGATTACAGTTCTTCTTATCCTGATCATGAAGATGAAAGGGAAGATATGAATGTTGATGAAGAAATAGATGGTCCTACAACACTGGAAGGAGATGATTTCCAATTGGTGTTGCCATCTGGAGTTACTGTTGGTCATAGATCTTTGATGAGATACTACAAACAGAATTTGACACAGAGCAGTCAAGCACTAGTAAAAAAATCTGATAGGAAACTGCACAGGCTTTTAGGTGTTTATAAAGCCCTGGGATGGGCGCCAAAGGAACAGAAAATGGCTGCCAAGAAAGCTCGGGATATACACTTCATGAAGCGTGTACAAGCAAAATGGGAGATGAAGATGTCCATGAAAACCAACAAGTTCCAGAAACATTACCGACCACAAGTGAATTTCTAG
Protein
MTMFSCITCKVLFNTSELQREHYKLDWHRYNLKRKVASIEPVTLEEFEERAKEHRESQNEKQDDSQYCQCCSKLFSTKNSYNNHLNSKKHKVSVERYTESQKDQENSGQSDTDSFVKVECTTGLTNERSKFVVVNTTDSGEDIETDSEIEELDSDEWDECRIQESDSLIKPRDCLFCVHHSKNMVKNLKHMSEVHSFFIPDVEFCINIRGLLLYLGEKISQGYMCLWCNDTGRTFYSMEAARAHMIDKGHCKMLHEGLALAEYADYYDYSSSYPDHEDEREDMNVDEEIDGPTTLEGDDFQLVLPSGVTVGHRSLMRYYKQNLTQSSQALVKKSDRKLHRLLGVYKALGWAPKEQKMAAKKARDIHFMKRVQAKWEMKMSMKTNKFQKHYRPQVNF
Summary
Uniprot
H9JE70
F6MDW8
A0A2A4K758
A0A0N1IQ74
S4PMS6
A0A0N0PE53
+ More
A0A194PKL0 A0A437BIY9 A0A1E1WBD2 A0A336LMG6 A0A336MPX7 A0A0K8TMN5 D2A0Y5 A0A182U4W7 Q7PYV1 A0A182HZU5 A0A182N691 A0A2J7PWA1 W5J939 B4JK00 A0A084W3I0 A0A2M4AG09 A0A182RNW7 W8CBD8 A0A182VCT9 A0A0M8ZVP9 A0A182PND0 A0A2M3Z8C2 A0A2M4AH64 A0A182JLC3 A0A1Q3F8P0 A0A182W009 A0A034WFP5 A0A067R4Z1 A0A0M3QZG4 B0WTB0 A0A182FR98 A0A1A9ZVD0 A0A1I8P8T1 A0A1Y1MEG7 A0A1Y1MCY0 D3TNE6 B3MS65 B4R6Q1 A0A2M4BR72 A0A1B0ATB6 A0A2P8YGS9 B4I6K9 B4L6D1 A0A1A9X4N0 B4MAD7 A0A182KCF9 A0A0A1WWC1 A0A182MC11 A0A2M4BQS9 A0A0K8V6I7 A0A1A9XAI9 A0A088AP54 A0A2M4BQX8 Q17FL5 Q9VX08 A0A1A9UPI1 A0A0L0BNE8 A0A182Q4F8 A0A3B0JY84 A0A1W4UNM1 A0A0P4W4M0 T1HLC3 A0A182Y456 B4Q2L5 B3NWY8 T1P9K8 A0A182H904 T1DSZ3 A0A182H0T8 B4NCQ8 A0A0T6B150 A0A182XGI1 A0A232FM87 A0A1B0G452 A0A2A3ER63 A0A1B6EGT6 U5ESK9 A0A1B6HB81 A0A1B6KJ29 J3JXA6 A0A0L7QRF7 A0A0A9ZIZ0 E0VLG5 R4WRM4 A0A0V0G6R1 A0A069DTG3 A0A182LQW6 A0A3Q0JMS3 A0A1B6FBF2 A0A0L8GDZ9 A0A224XJ09 R7V521 A0A1W4X3P0
A0A194PKL0 A0A437BIY9 A0A1E1WBD2 A0A336LMG6 A0A336MPX7 A0A0K8TMN5 D2A0Y5 A0A182U4W7 Q7PYV1 A0A182HZU5 A0A182N691 A0A2J7PWA1 W5J939 B4JK00 A0A084W3I0 A0A2M4AG09 A0A182RNW7 W8CBD8 A0A182VCT9 A0A0M8ZVP9 A0A182PND0 A0A2M3Z8C2 A0A2M4AH64 A0A182JLC3 A0A1Q3F8P0 A0A182W009 A0A034WFP5 A0A067R4Z1 A0A0M3QZG4 B0WTB0 A0A182FR98 A0A1A9ZVD0 A0A1I8P8T1 A0A1Y1MEG7 A0A1Y1MCY0 D3TNE6 B3MS65 B4R6Q1 A0A2M4BR72 A0A1B0ATB6 A0A2P8YGS9 B4I6K9 B4L6D1 A0A1A9X4N0 B4MAD7 A0A182KCF9 A0A0A1WWC1 A0A182MC11 A0A2M4BQS9 A0A0K8V6I7 A0A1A9XAI9 A0A088AP54 A0A2M4BQX8 Q17FL5 Q9VX08 A0A1A9UPI1 A0A0L0BNE8 A0A182Q4F8 A0A3B0JY84 A0A1W4UNM1 A0A0P4W4M0 T1HLC3 A0A182Y456 B4Q2L5 B3NWY8 T1P9K8 A0A182H904 T1DSZ3 A0A182H0T8 B4NCQ8 A0A0T6B150 A0A182XGI1 A0A232FM87 A0A1B0G452 A0A2A3ER63 A0A1B6EGT6 U5ESK9 A0A1B6HB81 A0A1B6KJ29 J3JXA6 A0A0L7QRF7 A0A0A9ZIZ0 E0VLG5 R4WRM4 A0A0V0G6R1 A0A069DTG3 A0A182LQW6 A0A3Q0JMS3 A0A1B6FBF2 A0A0L8GDZ9 A0A224XJ09 R7V521 A0A1W4X3P0
Pubmed
19121390
26354079
23622113
26369729
18362917
19820115
+ More
12364791 14747013 17210077 20920257 23761445 17994087 24438588 24495485 25348373 24845553 28004739 20353571 29403074 25830018 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 27129103 25244985 17550304 25315136 26483478 28648823 22516182 23537049 25401762 26823975 20566863 23691247 26334808 20966253 23254933
12364791 14747013 17210077 20920257 23761445 17994087 24438588 24495485 25348373 24845553 28004739 20353571 29403074 25830018 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 27129103 25244985 17550304 25315136 26483478 28648823 22516182 23537049 25401762 26823975 20566863 23691247 26334808 20966253 23254933
EMBL
BABH01002657
BABH01002658
JF793648
AEF58997.1
NWSH01000058
PCG80077.1
+ More
KQ459937 KPJ19226.1 GAIX01000021 JAA92539.1 KPJ19225.1 KQ459602 KPI93269.1 RSAL01000049 RVE50407.1 GDQN01007129 GDQN01006746 JAT83925.1 JAT84308.1 UFQT01000056 SSX19110.1 UFQS01002082 UFQT01002082 SSX13165.1 SSX32604.1 GDAI01002170 JAI15433.1 KQ971338 EFA01610.1 AAAB01008987 EAA00968.2 APCN01002013 NEVH01020936 PNF20600.1 ADMH02001839 ETN60967.1 CH916370 EDV99902.1 ATLV01020030 KE525292 KFB44774.1 GGFK01006331 MBW39652.1 GAMC01000574 JAC05982.1 KQ435859 KOX70573.1 GGFM01004028 MBW24779.1 GGFK01006756 MBW40077.1 GFDL01011162 JAV23883.1 GAKP01005433 JAC53519.1 KK852969 KDR13107.1 CP012528 ALC49329.1 DS232083 EDS34338.1 GEZM01038208 JAV81717.1 GEZM01038209 GEZM01038207 JAV81716.1 EZ422948 ADD19224.1 CH902622 EDV34620.1 CM000366 EDX18250.1 GGFJ01006280 MBW55421.1 JXJN01003232 PYGN01000605 PSN43459.1 CH480823 EDW55957.1 CH933812 EDW05927.1 CH940655 EDW66196.1 GBXI01011120 JAD03172.1 AXCM01004114 GGFJ01006279 MBW55420.1 GDHF01017966 JAI34348.1 GGFJ01006281 MBW55422.1 CH477270 EAT45328.1 AE014298 AY061122 AAF48775.1 AAL28670.1 JRES01001596 KNC21625.1 AXCN02000273 OUUW01000003 SPP77691.1 GDKW01000308 JAI56287.1 ACPB03017679 CM000162 EDX02656.1 CH954180 EDV46535.1 KA645412 AFP60041.1 JXUM01119701 KQ566317 KXJ70241.1 GAMD01001269 JAB00322.1 JXUM01101940 KQ564686 KXJ71900.1 CH964239 EDW82617.1 LJIG01022448 KRT80551.1 NNAY01000042 OXU31619.1 CCAG010008295 KZ288193 PBC33994.1 GEDC01013931 GEDC01000198 JAS23367.1 JAS37100.1 GANO01003204 JAB56667.1 GECU01035750 JAS71956.1 GEBQ01028524 JAT11453.1 APGK01046986 BT127875 KB741073 KB632433 AEE62837.1 ENN74382.1 ERL95557.1 KQ414778 KOC61220.1 GBHO01000434 GBRD01004845 GDHC01002179 JAG43170.1 JAG60976.1 JAQ16450.1 DS235271 EEB14221.1 AK417342 BAN20557.1 GECL01002329 JAP03795.1 GBGD01001838 JAC87051.1 GECZ01022298 JAS47471.1 KQ422493 KOF74770.1 GFTR01005392 JAW11034.1 AMQN01004984 KB294881 ELU13958.1
KQ459937 KPJ19226.1 GAIX01000021 JAA92539.1 KPJ19225.1 KQ459602 KPI93269.1 RSAL01000049 RVE50407.1 GDQN01007129 GDQN01006746 JAT83925.1 JAT84308.1 UFQT01000056 SSX19110.1 UFQS01002082 UFQT01002082 SSX13165.1 SSX32604.1 GDAI01002170 JAI15433.1 KQ971338 EFA01610.1 AAAB01008987 EAA00968.2 APCN01002013 NEVH01020936 PNF20600.1 ADMH02001839 ETN60967.1 CH916370 EDV99902.1 ATLV01020030 KE525292 KFB44774.1 GGFK01006331 MBW39652.1 GAMC01000574 JAC05982.1 KQ435859 KOX70573.1 GGFM01004028 MBW24779.1 GGFK01006756 MBW40077.1 GFDL01011162 JAV23883.1 GAKP01005433 JAC53519.1 KK852969 KDR13107.1 CP012528 ALC49329.1 DS232083 EDS34338.1 GEZM01038208 JAV81717.1 GEZM01038209 GEZM01038207 JAV81716.1 EZ422948 ADD19224.1 CH902622 EDV34620.1 CM000366 EDX18250.1 GGFJ01006280 MBW55421.1 JXJN01003232 PYGN01000605 PSN43459.1 CH480823 EDW55957.1 CH933812 EDW05927.1 CH940655 EDW66196.1 GBXI01011120 JAD03172.1 AXCM01004114 GGFJ01006279 MBW55420.1 GDHF01017966 JAI34348.1 GGFJ01006281 MBW55422.1 CH477270 EAT45328.1 AE014298 AY061122 AAF48775.1 AAL28670.1 JRES01001596 KNC21625.1 AXCN02000273 OUUW01000003 SPP77691.1 GDKW01000308 JAI56287.1 ACPB03017679 CM000162 EDX02656.1 CH954180 EDV46535.1 KA645412 AFP60041.1 JXUM01119701 KQ566317 KXJ70241.1 GAMD01001269 JAB00322.1 JXUM01101940 KQ564686 KXJ71900.1 CH964239 EDW82617.1 LJIG01022448 KRT80551.1 NNAY01000042 OXU31619.1 CCAG010008295 KZ288193 PBC33994.1 GEDC01013931 GEDC01000198 JAS23367.1 JAS37100.1 GANO01003204 JAB56667.1 GECU01035750 JAS71956.1 GEBQ01028524 JAT11453.1 APGK01046986 BT127875 KB741073 KB632433 AEE62837.1 ENN74382.1 ERL95557.1 KQ414778 KOC61220.1 GBHO01000434 GBRD01004845 GDHC01002179 JAG43170.1 JAG60976.1 JAQ16450.1 DS235271 EEB14221.1 AK417342 BAN20557.1 GECL01002329 JAP03795.1 GBGD01001838 JAC87051.1 GECZ01022298 JAS47471.1 KQ422493 KOF74770.1 GFTR01005392 JAW11034.1 AMQN01004984 KB294881 ELU13958.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007266
+ More
UP000075902 UP000007062 UP000075840 UP000075884 UP000235965 UP000000673 UP000001070 UP000030765 UP000075900 UP000075903 UP000053105 UP000075885 UP000075880 UP000075920 UP000027135 UP000092553 UP000002320 UP000069272 UP000092445 UP000095300 UP000007801 UP000000304 UP000092460 UP000245037 UP000001292 UP000009192 UP000091820 UP000008792 UP000075881 UP000075883 UP000092443 UP000005203 UP000008820 UP000000803 UP000078200 UP000037069 UP000075886 UP000268350 UP000192221 UP000015103 UP000076408 UP000002282 UP000008711 UP000095301 UP000069940 UP000249989 UP000007798 UP000076407 UP000215335 UP000092444 UP000242457 UP000019118 UP000030742 UP000053825 UP000009046 UP000075882 UP000079169 UP000053454 UP000014760 UP000192223
UP000075902 UP000007062 UP000075840 UP000075884 UP000235965 UP000000673 UP000001070 UP000030765 UP000075900 UP000075903 UP000053105 UP000075885 UP000075880 UP000075920 UP000027135 UP000092553 UP000002320 UP000069272 UP000092445 UP000095300 UP000007801 UP000000304 UP000092460 UP000245037 UP000001292 UP000009192 UP000091820 UP000008792 UP000075881 UP000075883 UP000092443 UP000005203 UP000008820 UP000000803 UP000078200 UP000037069 UP000075886 UP000268350 UP000192221 UP000015103 UP000076408 UP000002282 UP000008711 UP000095301 UP000069940 UP000249989 UP000007798 UP000076407 UP000215335 UP000092444 UP000242457 UP000019118 UP000030742 UP000053825 UP000009046 UP000075882 UP000079169 UP000053454 UP000014760 UP000192223
Interpro
SUPFAM
SSF57667
SSF57667
ProteinModelPortal
H9JE70
F6MDW8
A0A2A4K758
A0A0N1IQ74
S4PMS6
A0A0N0PE53
+ More
A0A194PKL0 A0A437BIY9 A0A1E1WBD2 A0A336LMG6 A0A336MPX7 A0A0K8TMN5 D2A0Y5 A0A182U4W7 Q7PYV1 A0A182HZU5 A0A182N691 A0A2J7PWA1 W5J939 B4JK00 A0A084W3I0 A0A2M4AG09 A0A182RNW7 W8CBD8 A0A182VCT9 A0A0M8ZVP9 A0A182PND0 A0A2M3Z8C2 A0A2M4AH64 A0A182JLC3 A0A1Q3F8P0 A0A182W009 A0A034WFP5 A0A067R4Z1 A0A0M3QZG4 B0WTB0 A0A182FR98 A0A1A9ZVD0 A0A1I8P8T1 A0A1Y1MEG7 A0A1Y1MCY0 D3TNE6 B3MS65 B4R6Q1 A0A2M4BR72 A0A1B0ATB6 A0A2P8YGS9 B4I6K9 B4L6D1 A0A1A9X4N0 B4MAD7 A0A182KCF9 A0A0A1WWC1 A0A182MC11 A0A2M4BQS9 A0A0K8V6I7 A0A1A9XAI9 A0A088AP54 A0A2M4BQX8 Q17FL5 Q9VX08 A0A1A9UPI1 A0A0L0BNE8 A0A182Q4F8 A0A3B0JY84 A0A1W4UNM1 A0A0P4W4M0 T1HLC3 A0A182Y456 B4Q2L5 B3NWY8 T1P9K8 A0A182H904 T1DSZ3 A0A182H0T8 B4NCQ8 A0A0T6B150 A0A182XGI1 A0A232FM87 A0A1B0G452 A0A2A3ER63 A0A1B6EGT6 U5ESK9 A0A1B6HB81 A0A1B6KJ29 J3JXA6 A0A0L7QRF7 A0A0A9ZIZ0 E0VLG5 R4WRM4 A0A0V0G6R1 A0A069DTG3 A0A182LQW6 A0A3Q0JMS3 A0A1B6FBF2 A0A0L8GDZ9 A0A224XJ09 R7V521 A0A1W4X3P0
A0A194PKL0 A0A437BIY9 A0A1E1WBD2 A0A336LMG6 A0A336MPX7 A0A0K8TMN5 D2A0Y5 A0A182U4W7 Q7PYV1 A0A182HZU5 A0A182N691 A0A2J7PWA1 W5J939 B4JK00 A0A084W3I0 A0A2M4AG09 A0A182RNW7 W8CBD8 A0A182VCT9 A0A0M8ZVP9 A0A182PND0 A0A2M3Z8C2 A0A2M4AH64 A0A182JLC3 A0A1Q3F8P0 A0A182W009 A0A034WFP5 A0A067R4Z1 A0A0M3QZG4 B0WTB0 A0A182FR98 A0A1A9ZVD0 A0A1I8P8T1 A0A1Y1MEG7 A0A1Y1MCY0 D3TNE6 B3MS65 B4R6Q1 A0A2M4BR72 A0A1B0ATB6 A0A2P8YGS9 B4I6K9 B4L6D1 A0A1A9X4N0 B4MAD7 A0A182KCF9 A0A0A1WWC1 A0A182MC11 A0A2M4BQS9 A0A0K8V6I7 A0A1A9XAI9 A0A088AP54 A0A2M4BQX8 Q17FL5 Q9VX08 A0A1A9UPI1 A0A0L0BNE8 A0A182Q4F8 A0A3B0JY84 A0A1W4UNM1 A0A0P4W4M0 T1HLC3 A0A182Y456 B4Q2L5 B3NWY8 T1P9K8 A0A182H904 T1DSZ3 A0A182H0T8 B4NCQ8 A0A0T6B150 A0A182XGI1 A0A232FM87 A0A1B0G452 A0A2A3ER63 A0A1B6EGT6 U5ESK9 A0A1B6HB81 A0A1B6KJ29 J3JXA6 A0A0L7QRF7 A0A0A9ZIZ0 E0VLG5 R4WRM4 A0A0V0G6R1 A0A069DTG3 A0A182LQW6 A0A3Q0JMS3 A0A1B6FBF2 A0A0L8GDZ9 A0A224XJ09 R7V521 A0A1W4X3P0
PDB
5APN
E-value=2.81342e-19,
Score=234
Ontologies
GO
PANTHER
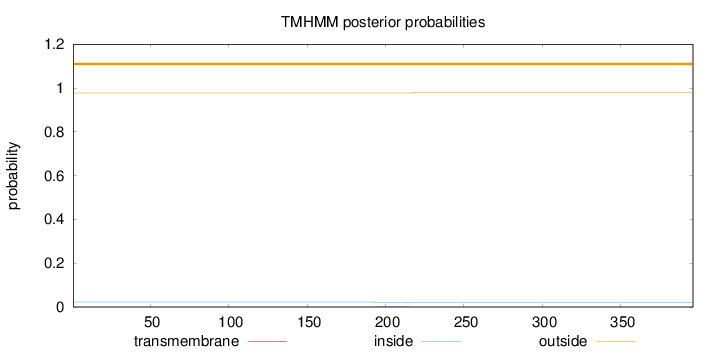
Topology
Length:
396
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0363300000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02258
outside
1 - 396
Population Genetic Test Statistics
Pi
246.854443
Theta
192.030196
Tajima's D
0.753926
CLR
0.380651
CSRT
0.588420578971051
Interpretation
Uncertain
