Gene
KWMTBOMO08910
Pre Gene Modal
BGIBMGA007715
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_LRSAM1-like_[Bombyx_mori]
Full name
Metalloendopeptidase
Location in the cell
Nuclear Reliability : 3.095
Sequence
CDS
ATGGGTTGTGCTGCATCTTGCATACAGGAAGCGACTATGTCGTTGTTCGGGAGATCTTCAAACAATATAGCTGACGATGATAGAGCTAAATTGGAAAGGAAGCTGTATGTTGCCAAAGAGTCCCCGGAGCCCGAATTCGACCTTTCTGATTGTAATTTGCGACAAATACCGTCCGGCACGTATTCTATTTGCAAGGTCTACAGAAAAGAATATATATATTTACACAAAAACGATTTACATTCCTTGGAAGACGGCGGCCAGTTGTCTGATTTGTATCTTATTAAAGTGCTCAACATTAGTTCTAACAAATTTTCTCATCTTCCCAGTGACATAAAATATTTAGTTAATGTTACTGAGTTGTATGTTAGTGATAATATGCTAAAGAATATAACAGACAATATTCAATACATGGAGAACTTGCAAGTTTTGGATGTTTCGAATAATAAACTAAAAAGTCTTACACCAGTTCTTGGCAAATTGAAATATTTAAGAAAATTGAACATTACTGGTAACAGAGAATTAACAGAAATATGTCCTGAATTATGTTTTGCAAGGAATTTGATTTGCTTGGAATTGGATATGGAATATATTACTTTTCCTCCAGTTGAGATTTGTCAGCAGGGAACAAGAGAAATAATGAGGTATTTATGTAATAAGATGAATGTGGAATATTCCCCAGCTGCTGAAGAAATTGAAGTGTTTTCGATTAGAACATCTAATTCTCTATTGGATCCGTTTGCTAAAGCACATTCTACTACTTGGGAAGAACAAGAAGCTTCTATTGTCGCTCAAGAGAACAAGATACACAAAATAGCTAAAGAACAAAGAGAAAAATTCTTGTCAAAGGTTCTACAAGAACAATTAGAGTTAGACTCTGAAATAGCTAAAGTTCATGAACAGAGGGAGACAGAAAGACAAAGACTGATTAAGGCCATTCAAGATGATGAAAAAGAAATAGAATGTTTGGTTAATAATTTCATTCAATTTGAATACTTGAAACCAGAAATAATTCAGCAACAGCTTGCTCATGAACAGGCAGAGCATGAAAGGCTGCTTGAAATAACCAAACAAAATTATGATAATGTAAAGAAATCTGAGATTTTGAAAGCAATGGAAATGCTCCTGGAGGAAGACAGGTCAATACAATATTCTAAAAAACATTATGAAGACTCTTTGAATAATATTAAGCAGAGCATGTTGATGCAAGATGCAGAAATAGCTGATAAAGTAACAGATTTAATTAAAGCTAAAGATCAATCAAGGACAGTATTAGTGCAACAGTTTCTTGAAGATCAAGATATCCAAAAAGCTATTGTTGCCTCTTTGCTGGAAAGGGTGGATGCTAGAAGTTGGAGTTTGAATCAAGAAATATCCTTGATCTCATCTCACCTTGCCAGGCTGAGTGTTATAGAACAAGAGAAGAAGAAATTACATATAGCATATAACTACAATGAGTTGCTAAACCAGAGAGTGCAACTTGTGAACTTTTTGGATGATCTCTTCAATCAACAAAATAAACGGAGGAAGCAGCTAATTCAAACTGTAAAAGAAATGGAAAATGAAAAAAGCCAAACGAATGACTTTTGGTTGAAAAACTACCAAAAGTTGATAGAATCTGCCCCAAAGACTTTGTTAGATGTTGGTAAATGTTTAGATCCAGTTTTTGCTAACTACTTGCTACAAGAAGGTGTTATACACTGTTTACCATTTTTGGTTAAATTTTTGTTTTCTGGTGATGTATTAAGTAATATAACATTGGAAAAATTGAAAGAGAGTGGAGTTTCACTTTCAGCTGACAGAGATGCAATTATGAGAGCAATCAACTTTTACATTGGTATCAAAAATTCTGAAAATTCTAATGCTAATTATGAACAACCAACATCTATACAACCAAGTGCTCCAATAGAGGATGTCGATGAGAACAATTGTACTGGAGTTGTAAATGTAGTTGAAGAATCTACTCTGGAATCGGAGTGCGTGATTTGTATGGATGCAAAATGTGAGGTGGTGTTCATACCATGTGGTCACATGTGTTGCTGTAAAGATTGCGGGGGAGGTGTTGAAGATTGTCCCATGTGCAGATGTAGTATTGAAAGGACAATCAAAGTTATGATGGCATGA
Protein
MGCAASCIQEATMSLFGRSSNNIADDDRAKLERKLYVAKESPEPEFDLSDCNLRQIPSGTYSICKVYRKEYIYLHKNDLHSLEDGGQLSDLYLIKVLNISSNKFSHLPSDIKYLVNVTELYVSDNMLKNITDNIQYMENLQVLDVSNNKLKSLTPVLGKLKYLRKLNITGNRELTEICPELCFARNLICLELDMEYITFPPVEICQQGTREIMRYLCNKMNVEYSPAAEEIEVFSIRTSNSLLDPFAKAHSTTWEEQEASIVAQENKIHKIAKEQREKFLSKVLQEQLELDSEIAKVHEQRETERQRLIKAIQDDEKEIECLVNNFIQFEYLKPEIIQQQLAHEQAEHERLLEITKQNYDNVKKSEILKAMEMLLEEDRSIQYSKKHYEDSLNNIKQSMLMQDAEIADKVTDLIKAKDQSRTVLVQQFLEDQDIQKAIVASLLERVDARSWSLNQEISLISSHLARLSVIEQEKKKLHIAYNYNELLNQRVQLVNFLDDLFNQQNKRRKQLIQTVKEMENEKSQTNDFWLKNYQKLIESAPKTLLDVGKCLDPVFANYLLQEGVIHCLPFLVKFLFSGDVLSNITLEKLKESGVSLSADRDAIMRAINFYIGIKNSENSNANYEQPTSIQPSAPIEDVDENNCTGVVNVVEESTLESECVICMDAKCEVVFIPCGHMCCCKDCGGGVEDCPMCRCSIERTIKVMMA
Summary
Cofactor
Zn(2+)
Feature
chain Metalloendopeptidase
Uniprot
H9JDW8
A0A2A4K781
A0A2H1V5S7
A0A1E1VXQ4
A0A0L7LKX1
A0A194PIW3
+ More
A0A212F3Y0 A0A2P8XNB1 A0A2J7RK73 A0A1Y1LHG4 E2BSU5 A0A1Y1LNA9 A0A067RJL5 A0A088A5Q9 A0A2A3E4Q0 A0A2J7RK67 U4UT61 A0A154PLJ7 A0A151WQV9 A0A0L7R9E4 A0A0C9PZK4 A0A336LWP4 A0A0N0BEG9 A0A1J1J9J4 Q17KA2 A0A1S4EZP6 A0A310S8Q3 A0A182YJ18 A0A1B6CD26 K7J6B0 A0A1Q3G3E3 A0A1Q3G387 A0A182NZR0 A0A1S4GNP6 A0A023EWL2 A0A2M3Z643 A0A2M3Z5Q8 A0A2M3Z5L5 A0A2M4AK24 A0A2M4BFN8 A0A2M4BF44 A0A2M4BF14 A0A195D275 W5J5R8 A0A232F1G5 A0A182KB84 A0A182FHU8 A0A2J7RK65 A0A084WCU2 A0A1Y1LHE3 A0A0C9Q9M5 A0A182RUX6 A0A1B0CHP9 A0A1Y1LJK6 A0NDS3 A0A182X0X0 A0A146MIF4 A0A182GZL1 A0A1L8DSN6 A0A1L8DSD9 A0A3M6UJZ9 A0A182H125 A0A146M263 A0A151I4H0 A0A1B0D9J5 A0A182J3G8 A0A182LB53 A0A182WMB3 A0A0L8IDQ0 A0A1B6L182 A0A182NLW7 A0A182R0I0 A0A182MCY2 A0A224XL14 A0A087UUE2 T1J7S0 A0A158NZC4 A0A369S958 A0A2R5LHF9 A0A182HPD8 F4W556 W5KZX0 A0A182VC69 A0A2L2YB68 A0A2L2YB49 A0A1L8F0P8 A0A131XRS5 A0A147BHV3 A0A210QV31 A0A293MSP1 A0A1L8F5Q9 F6Q0X6
A0A212F3Y0 A0A2P8XNB1 A0A2J7RK73 A0A1Y1LHG4 E2BSU5 A0A1Y1LNA9 A0A067RJL5 A0A088A5Q9 A0A2A3E4Q0 A0A2J7RK67 U4UT61 A0A154PLJ7 A0A151WQV9 A0A0L7R9E4 A0A0C9PZK4 A0A336LWP4 A0A0N0BEG9 A0A1J1J9J4 Q17KA2 A0A1S4EZP6 A0A310S8Q3 A0A182YJ18 A0A1B6CD26 K7J6B0 A0A1Q3G3E3 A0A1Q3G387 A0A182NZR0 A0A1S4GNP6 A0A023EWL2 A0A2M3Z643 A0A2M3Z5Q8 A0A2M3Z5L5 A0A2M4AK24 A0A2M4BFN8 A0A2M4BF44 A0A2M4BF14 A0A195D275 W5J5R8 A0A232F1G5 A0A182KB84 A0A182FHU8 A0A2J7RK65 A0A084WCU2 A0A1Y1LHE3 A0A0C9Q9M5 A0A182RUX6 A0A1B0CHP9 A0A1Y1LJK6 A0NDS3 A0A182X0X0 A0A146MIF4 A0A182GZL1 A0A1L8DSN6 A0A1L8DSD9 A0A3M6UJZ9 A0A182H125 A0A146M263 A0A151I4H0 A0A1B0D9J5 A0A182J3G8 A0A182LB53 A0A182WMB3 A0A0L8IDQ0 A0A1B6L182 A0A182NLW7 A0A182R0I0 A0A182MCY2 A0A224XL14 A0A087UUE2 T1J7S0 A0A158NZC4 A0A369S958 A0A2R5LHF9 A0A182HPD8 F4W556 W5KZX0 A0A182VC69 A0A2L2YB68 A0A2L2YB49 A0A1L8F0P8 A0A131XRS5 A0A147BHV3 A0A210QV31 A0A293MSP1 A0A1L8F5Q9 F6Q0X6
EC Number
3.4.24.-
Pubmed
EMBL
BABH01002692
NWSH01000058
PCG80091.1
ODYU01000638
SOQ35752.1
GDQN01011554
+ More
JAT79500.1 JTDY01000792 KOB75856.1 KQ459602 KPI93262.1 AGBW02010474 OWR48440.1 PYGN01001664 PSN33497.1 NEVH01002981 PNF41232.1 GEZM01055241 JAV73073.1 GL450269 EFN81237.1 GEZM01055239 JAV73076.1 KK852591 KDR20711.1 KZ288400 PBC26262.1 PNF41227.1 KB632353 ERL93341.1 KQ434972 KZC12745.1 KQ982821 KYQ50187.1 KQ414627 KOC67473.1 GBYB01006988 JAG76755.1 UFQS01000246 UFQT01000246 SSX01968.1 SSX22345.1 KQ435828 KOX71892.1 CVRI01000075 CRL08548.1 CH477227 EAT47083.1 KQ769940 OAD52731.1 GEDC01029170 GEDC01026033 JAS08128.1 JAS11265.1 GFDL01000759 JAV34286.1 GFDL01000782 JAV34263.1 AAAB01008900 GBBI01004992 JAC13720.1 GGFM01003167 MBW23918.1 GGFM01003106 MBW23857.1 GGFM01003055 MBW23806.1 GGFK01007761 MBW41082.1 GGFJ01002497 MBW51638.1 GGFJ01002496 MBW51637.1 GGFJ01002498 MBW51639.1 KQ977004 KYN06489.1 ADMH02002165 ETN58119.1 NNAY01001263 OXU24565.1 PNF41233.1 ATLV01022885 KE525337 KFB48036.1 GEZM01055242 JAV73072.1 GBYB01011078 JAG80845.1 AJWK01012578 GEZM01055243 JAV73071.1 EAU76810.2 GDHC01000179 JAQ18450.1 JXUM01099713 KQ564513 KXJ72150.1 GFDF01004687 JAV09397.1 GFDF01004686 JAV09398.1 RCHS01001361 RMX53945.1 JXUM01102357 KQ564721 KXJ71849.1 GDHC01012241 GDHC01005294 GDHC01004857 JAQ06388.1 JAQ13335.1 JAQ13772.1 KQ976448 KYM85875.1 AJVK01012976 KQ415932 KOF99602.1 GEBQ01022508 JAT17469.1 AXCN02000947 AXCM01004675 GFTR01007707 JAW08719.1 KK121665 KFM80981.1 JH431939 ADTU01004410 ADTU01004411 ADTU01004412 NOWV01000038 RDD43445.1 GGLE01004834 MBY08960.1 APCN01002982 GL887605 EGI70652.1 IAAA01015481 LAA05396.1 IAAA01015480 LAA05392.1 CM004481 OCT65204.1 GEFM01006931 JAP68865.1 GEGO01005529 JAR89875.1 NEDP02001728 OWF52585.1 GFWV01017457 MAA42185.1 CM004480 OCT66915.1 EAAA01002990
JAT79500.1 JTDY01000792 KOB75856.1 KQ459602 KPI93262.1 AGBW02010474 OWR48440.1 PYGN01001664 PSN33497.1 NEVH01002981 PNF41232.1 GEZM01055241 JAV73073.1 GL450269 EFN81237.1 GEZM01055239 JAV73076.1 KK852591 KDR20711.1 KZ288400 PBC26262.1 PNF41227.1 KB632353 ERL93341.1 KQ434972 KZC12745.1 KQ982821 KYQ50187.1 KQ414627 KOC67473.1 GBYB01006988 JAG76755.1 UFQS01000246 UFQT01000246 SSX01968.1 SSX22345.1 KQ435828 KOX71892.1 CVRI01000075 CRL08548.1 CH477227 EAT47083.1 KQ769940 OAD52731.1 GEDC01029170 GEDC01026033 JAS08128.1 JAS11265.1 GFDL01000759 JAV34286.1 GFDL01000782 JAV34263.1 AAAB01008900 GBBI01004992 JAC13720.1 GGFM01003167 MBW23918.1 GGFM01003106 MBW23857.1 GGFM01003055 MBW23806.1 GGFK01007761 MBW41082.1 GGFJ01002497 MBW51638.1 GGFJ01002496 MBW51637.1 GGFJ01002498 MBW51639.1 KQ977004 KYN06489.1 ADMH02002165 ETN58119.1 NNAY01001263 OXU24565.1 PNF41233.1 ATLV01022885 KE525337 KFB48036.1 GEZM01055242 JAV73072.1 GBYB01011078 JAG80845.1 AJWK01012578 GEZM01055243 JAV73071.1 EAU76810.2 GDHC01000179 JAQ18450.1 JXUM01099713 KQ564513 KXJ72150.1 GFDF01004687 JAV09397.1 GFDF01004686 JAV09398.1 RCHS01001361 RMX53945.1 JXUM01102357 KQ564721 KXJ71849.1 GDHC01012241 GDHC01005294 GDHC01004857 JAQ06388.1 JAQ13335.1 JAQ13772.1 KQ976448 KYM85875.1 AJVK01012976 KQ415932 KOF99602.1 GEBQ01022508 JAT17469.1 AXCN02000947 AXCM01004675 GFTR01007707 JAW08719.1 KK121665 KFM80981.1 JH431939 ADTU01004410 ADTU01004411 ADTU01004412 NOWV01000038 RDD43445.1 GGLE01004834 MBY08960.1 APCN01002982 GL887605 EGI70652.1 IAAA01015481 LAA05396.1 IAAA01015480 LAA05392.1 CM004481 OCT65204.1 GEFM01006931 JAP68865.1 GEGO01005529 JAR89875.1 NEDP02001728 OWF52585.1 GFWV01017457 MAA42185.1 CM004480 OCT66915.1 EAAA01002990
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000007151
UP000245037
+ More
UP000235965 UP000008237 UP000027135 UP000005203 UP000242457 UP000030742 UP000076502 UP000075809 UP000053825 UP000053105 UP000183832 UP000008820 UP000076408 UP000002358 UP000075885 UP000078542 UP000000673 UP000215335 UP000075881 UP000069272 UP000030765 UP000075900 UP000092461 UP000007062 UP000076407 UP000069940 UP000249989 UP000275408 UP000078540 UP000092462 UP000075880 UP000075882 UP000075920 UP000053454 UP000075884 UP000075886 UP000075883 UP000054359 UP000005205 UP000253843 UP000075840 UP000007755 UP000018467 UP000075903 UP000186698 UP000242188 UP000008144
UP000235965 UP000008237 UP000027135 UP000005203 UP000242457 UP000030742 UP000076502 UP000075809 UP000053825 UP000053105 UP000183832 UP000008820 UP000076408 UP000002358 UP000075885 UP000078542 UP000000673 UP000215335 UP000075881 UP000069272 UP000030765 UP000075900 UP000092461 UP000007062 UP000076407 UP000069940 UP000249989 UP000275408 UP000078540 UP000092462 UP000075880 UP000075882 UP000075920 UP000053454 UP000075884 UP000075886 UP000075883 UP000054359 UP000005205 UP000253843 UP000075840 UP000007755 UP000018467 UP000075903 UP000186698 UP000242188 UP000008144
PRIDE
Interpro
IPR001841
Znf_RING
+ More
IPR032675 LRR_dom_sf
IPR013083 Znf_RING/FYVE/PHD
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR013761 SAM/pointed_sf
IPR024079 MetalloPept_cat_dom_sf
IPR006026 Peptidase_Metallo
IPR034035 Astacin-like_dom
IPR001506 Peptidase_M12A
IPR001660 SAM
IPR025875 Leu-rich_rpt_4
IPR032675 LRR_dom_sf
IPR013083 Znf_RING/FYVE/PHD
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR013761 SAM/pointed_sf
IPR024079 MetalloPept_cat_dom_sf
IPR006026 Peptidase_Metallo
IPR034035 Astacin-like_dom
IPR001506 Peptidase_M12A
IPR001660 SAM
IPR025875 Leu-rich_rpt_4
SUPFAM
SSF47769
SSF47769
Gene 3D
ProteinModelPortal
H9JDW8
A0A2A4K781
A0A2H1V5S7
A0A1E1VXQ4
A0A0L7LKX1
A0A194PIW3
+ More
A0A212F3Y0 A0A2P8XNB1 A0A2J7RK73 A0A1Y1LHG4 E2BSU5 A0A1Y1LNA9 A0A067RJL5 A0A088A5Q9 A0A2A3E4Q0 A0A2J7RK67 U4UT61 A0A154PLJ7 A0A151WQV9 A0A0L7R9E4 A0A0C9PZK4 A0A336LWP4 A0A0N0BEG9 A0A1J1J9J4 Q17KA2 A0A1S4EZP6 A0A310S8Q3 A0A182YJ18 A0A1B6CD26 K7J6B0 A0A1Q3G3E3 A0A1Q3G387 A0A182NZR0 A0A1S4GNP6 A0A023EWL2 A0A2M3Z643 A0A2M3Z5Q8 A0A2M3Z5L5 A0A2M4AK24 A0A2M4BFN8 A0A2M4BF44 A0A2M4BF14 A0A195D275 W5J5R8 A0A232F1G5 A0A182KB84 A0A182FHU8 A0A2J7RK65 A0A084WCU2 A0A1Y1LHE3 A0A0C9Q9M5 A0A182RUX6 A0A1B0CHP9 A0A1Y1LJK6 A0NDS3 A0A182X0X0 A0A146MIF4 A0A182GZL1 A0A1L8DSN6 A0A1L8DSD9 A0A3M6UJZ9 A0A182H125 A0A146M263 A0A151I4H0 A0A1B0D9J5 A0A182J3G8 A0A182LB53 A0A182WMB3 A0A0L8IDQ0 A0A1B6L182 A0A182NLW7 A0A182R0I0 A0A182MCY2 A0A224XL14 A0A087UUE2 T1J7S0 A0A158NZC4 A0A369S958 A0A2R5LHF9 A0A182HPD8 F4W556 W5KZX0 A0A182VC69 A0A2L2YB68 A0A2L2YB49 A0A1L8F0P8 A0A131XRS5 A0A147BHV3 A0A210QV31 A0A293MSP1 A0A1L8F5Q9 F6Q0X6
A0A212F3Y0 A0A2P8XNB1 A0A2J7RK73 A0A1Y1LHG4 E2BSU5 A0A1Y1LNA9 A0A067RJL5 A0A088A5Q9 A0A2A3E4Q0 A0A2J7RK67 U4UT61 A0A154PLJ7 A0A151WQV9 A0A0L7R9E4 A0A0C9PZK4 A0A336LWP4 A0A0N0BEG9 A0A1J1J9J4 Q17KA2 A0A1S4EZP6 A0A310S8Q3 A0A182YJ18 A0A1B6CD26 K7J6B0 A0A1Q3G3E3 A0A1Q3G387 A0A182NZR0 A0A1S4GNP6 A0A023EWL2 A0A2M3Z643 A0A2M3Z5Q8 A0A2M3Z5L5 A0A2M4AK24 A0A2M4BFN8 A0A2M4BF44 A0A2M4BF14 A0A195D275 W5J5R8 A0A232F1G5 A0A182KB84 A0A182FHU8 A0A2J7RK65 A0A084WCU2 A0A1Y1LHE3 A0A0C9Q9M5 A0A182RUX6 A0A1B0CHP9 A0A1Y1LJK6 A0NDS3 A0A182X0X0 A0A146MIF4 A0A182GZL1 A0A1L8DSN6 A0A1L8DSD9 A0A3M6UJZ9 A0A182H125 A0A146M263 A0A151I4H0 A0A1B0D9J5 A0A182J3G8 A0A182LB53 A0A182WMB3 A0A0L8IDQ0 A0A1B6L182 A0A182NLW7 A0A182R0I0 A0A182MCY2 A0A224XL14 A0A087UUE2 T1J7S0 A0A158NZC4 A0A369S958 A0A2R5LHF9 A0A182HPD8 F4W556 W5KZX0 A0A182VC69 A0A2L2YB68 A0A2L2YB49 A0A1L8F0P8 A0A131XRS5 A0A147BHV3 A0A210QV31 A0A293MSP1 A0A1L8F5Q9 F6Q0X6
PDB
6DJB
E-value=4.87758e-11,
Score=166
Ontologies
GO
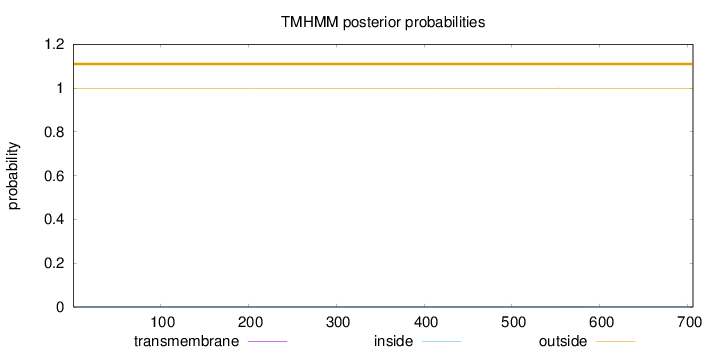
Topology
Length:
706
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02086
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00042
outside
1 - 706
Population Genetic Test Statistics
Pi
302.895923
Theta
211.295264
Tajima's D
1.301612
CLR
0
CSRT
0.737513124343783
Interpretation
Uncertain
