Gene
KWMTBOMO08907
Annotation
mariner_transposase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.47
Sequence
CDS
ATGGAATTGACTCGAGAAAATTCAAGAGCGATGATTTATTATGACTTTCGAAGTGGTTTAACACAAAAACAGTGTGTTGATCGGATGATTTCTGCATTTGGTGATGAAGCCCCATCCAAAACCACAATTGATTGCTGGTTTGCTGAGTTTCAACGTGGACGTGTCAAGCTCAGTGATGAACCCCGTCAAGGTCGTCAAAAAGCTGCAGTCACCCAAGAAAACGTTGATGCTGTCGTTTCTGAACTCCGTAAAGAGAACTGCAACCGCCGCATCATCCTCCATCACGACAATGCGAGTTCTCACACCGCGCACAGAACAAAAGAGTTTTTAGAGCAAGAAAACATAGAATTATTAGACCATCCGCCGTACAGCCCCGACCTAAGCCCTAATGATTTCTATACTTTCCCTAAAATAAAGAATAAATTGTGTGGACAGAGATTTTCATCACCTGAAGAAGCTGTGGACGCCTACAAAACGGCCATTTTGGAGACCCCAACTTCCGAATGGAATGGTTGCTTCAATGATTGGTTCCATCGTATGGAAAAATATGTCAAATTTCGCGGAGAATACTTCTAA
Protein
MELTRENSRAMIYYDFRSGLTQKQCVDRMISAFGDEAPSKTTIDCWFAEFQRGRVKLSDEPRQGRQKAAVTQENVDAVVSELRKENCNRRIILHHDNASSHTAHRTKEFLEQENIELLDHPPYSPDLSPNDFYTFPKIKNKLCGQRFSSPEEAVDAYKTAILETPTSEWNGCFNDWFHRMEKYVKFRGEYF
Summary
Uniprot
A0A195DZA6
F4WS09
A0A1B6G5C6
A0A034VN11
A0A0V0G5J0
A0A069DT60
+ More
A0A023F6H6 A0A1L5BY10 A0A267ESW3 A0A151WTD8 A0A2J7R478 A0A2J7QWK6 A0A2J7PW98 A0A2J7QP01 A0A2J7Q4S5 A0A0K2VK65 A0A151K0H9 A0A151K0I0 A0A151JXR4 A0A151JTI7 A0A151JZN7 A0A151JWC9 A0A151JWE8 A0A1W0WPR3 A0A2J7QND6 A0A151JUK9 A0A2J7RS37 A0A2J7R3C8 A0A151XI18 F4WPJ1 A0A151IRY2 A0A151JTQ3 A0A2J7PUI4 A0A2J7PJ78 A0A2J7R8T0 A0A151JU59 A0A195EWQ5 A0A151JAL8 B3CK30 A0A195EUP8 A0A195FM19 A0A2J7QJG3 A0A151WL87 A0A2J7PGR2 A0A2J7PRZ3 B3CK24 B3CK26 A0A026W2H3 B3CK19 A0A2J7QX91 A0A2J7QFV5 F4WVW5 A0A2J7PFM0 F4WY44 A0A2J7QVE3 A0A2J7Q656 A0A023F6W4 A0A2J7QQ11 A0A023F6P2 A0A2J7Q009 A0A2J7R0Z3 A0A2J7RQG1 A0A2J7PE56 F4X169 A0A2J7Q0F6 A0A2J7QR48
A0A023F6H6 A0A1L5BY10 A0A267ESW3 A0A151WTD8 A0A2J7R478 A0A2J7QWK6 A0A2J7PW98 A0A2J7QP01 A0A2J7Q4S5 A0A0K2VK65 A0A151K0H9 A0A151K0I0 A0A151JXR4 A0A151JTI7 A0A151JZN7 A0A151JWC9 A0A151JWE8 A0A1W0WPR3 A0A2J7QND6 A0A151JUK9 A0A2J7RS37 A0A2J7R3C8 A0A151XI18 F4WPJ1 A0A151IRY2 A0A151JTQ3 A0A2J7PUI4 A0A2J7PJ78 A0A2J7R8T0 A0A151JU59 A0A195EWQ5 A0A151JAL8 B3CK30 A0A195EUP8 A0A195FM19 A0A2J7QJG3 A0A151WL87 A0A2J7PGR2 A0A2J7PRZ3 B3CK24 B3CK26 A0A026W2H3 B3CK19 A0A2J7QX91 A0A2J7QFV5 F4WVW5 A0A2J7PFM0 F4WY44 A0A2J7QVE3 A0A2J7Q656 A0A023F6W4 A0A2J7QQ11 A0A023F6P2 A0A2J7Q009 A0A2J7R0Z3 A0A2J7RQG1 A0A2J7PE56 F4X169 A0A2J7Q0F6 A0A2J7QR48
EMBL
KQ980031
KYN18156.1
GL888295
EGI62988.1
GECZ01012256
JAS57513.1
+ More
GAKP01015435 JAC43517.1 GECL01002811 JAP03313.1 GBGD01001983 JAC86906.1 GBBI01001865 JAC16847.1 KX930998 APL98291.1 NIVC01001798 PAA63957.1 KQ982756 KYQ51169.1 NEVH01007794 PNF35638.1 NEVH01009765 PNF32944.1 NEVH01020936 PNF20612.1 NEVH01012098 PNF30316.1 NEVH01018377 PNF23576.1 HACA01033482 CDW50843.1 KQ981250 KYN44215.1 KYN44220.1 KQ981615 KQ981453 KQ981332 KYN39295.1 KYN41582.1 KYN42673.1 KQ981885 KYN33839.1 KYN33874.1 KQ981337 KYN42545.1 KQ981661 KYN38529.1 KQ981643 KYN38682.1 MTYJ01000064 OQV17200.1 NEVH01013196 PNF30090.1 KQ981733 KYN36550.1 NEVH01000595 PNF43644.1 NEVH01007822 PNF35315.1 KQ982126 KYQ59860.1 GL888247 EGI63876.1 KQ981102 KYN09459.1 KQ981832 KYN35065.1 NEVH01021198 NEVH01008206 NEVH01006571 PNF19983.1 PNF34678.1 PNF37800.1 NEVH01024950 NEVH01022640 NEVH01006723 PNF16374.1 PNF18508.1 PNF36951.1 NEVH01006721 PNF37233.1 KQ981816 KYN35367.1 KQ981948 KYN32581.1 KQ979265 KYN22061.1 AM906154 CAP20068.1 KQ981975 KYN31622.1 KQ981430 KYN41745.1 NEVH01013555 PNF28730.1 KQ982975 KYQ48580.1 NEVH01025167 PNF15526.1 NEVH01021944 PNF19091.1 AM906142 AM906144 CAP20056.1 AM906145 CAP20059.1 KK107532 EZA49234.1 AM906136 CAP20050.1 NEVH01009384 PNF33206.1 NEVH01014841 PNF27477.1 GL888401 EGI61662.1 NEVH01025655 PNF15130.1 GL888437 EGI60875.1 NEVH01010477 PNF32541.1 NEVH01017540 PNF24058.1 GBBI01001742 JAC16970.1 NEVH01012087 PNF30678.1 GBBI01001812 JAC16900.1 NEVH01020329 PNF21909.1 NEVH01008216 PNF34501.1 NEVH01001337 PNF43072.1 NEVH01026109 PNF14609.1 GL888527 EGI59805.1 NEVH01019969 PNF22065.1 NEVH01011986 PNF31047.1
GAKP01015435 JAC43517.1 GECL01002811 JAP03313.1 GBGD01001983 JAC86906.1 GBBI01001865 JAC16847.1 KX930998 APL98291.1 NIVC01001798 PAA63957.1 KQ982756 KYQ51169.1 NEVH01007794 PNF35638.1 NEVH01009765 PNF32944.1 NEVH01020936 PNF20612.1 NEVH01012098 PNF30316.1 NEVH01018377 PNF23576.1 HACA01033482 CDW50843.1 KQ981250 KYN44215.1 KYN44220.1 KQ981615 KQ981453 KQ981332 KYN39295.1 KYN41582.1 KYN42673.1 KQ981885 KYN33839.1 KYN33874.1 KQ981337 KYN42545.1 KQ981661 KYN38529.1 KQ981643 KYN38682.1 MTYJ01000064 OQV17200.1 NEVH01013196 PNF30090.1 KQ981733 KYN36550.1 NEVH01000595 PNF43644.1 NEVH01007822 PNF35315.1 KQ982126 KYQ59860.1 GL888247 EGI63876.1 KQ981102 KYN09459.1 KQ981832 KYN35065.1 NEVH01021198 NEVH01008206 NEVH01006571 PNF19983.1 PNF34678.1 PNF37800.1 NEVH01024950 NEVH01022640 NEVH01006723 PNF16374.1 PNF18508.1 PNF36951.1 NEVH01006721 PNF37233.1 KQ981816 KYN35367.1 KQ981948 KYN32581.1 KQ979265 KYN22061.1 AM906154 CAP20068.1 KQ981975 KYN31622.1 KQ981430 KYN41745.1 NEVH01013555 PNF28730.1 KQ982975 KYQ48580.1 NEVH01025167 PNF15526.1 NEVH01021944 PNF19091.1 AM906142 AM906144 CAP20056.1 AM906145 CAP20059.1 KK107532 EZA49234.1 AM906136 CAP20050.1 NEVH01009384 PNF33206.1 NEVH01014841 PNF27477.1 GL888401 EGI61662.1 NEVH01025655 PNF15130.1 GL888437 EGI60875.1 NEVH01010477 PNF32541.1 NEVH01017540 PNF24058.1 GBBI01001742 JAC16970.1 NEVH01012087 PNF30678.1 GBBI01001812 JAC16900.1 NEVH01020329 PNF21909.1 NEVH01008216 PNF34501.1 NEVH01001337 PNF43072.1 NEVH01026109 PNF14609.1 GL888527 EGI59805.1 NEVH01019969 PNF22065.1 NEVH01011986 PNF31047.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A195DZA6
F4WS09
A0A1B6G5C6
A0A034VN11
A0A0V0G5J0
A0A069DT60
+ More
A0A023F6H6 A0A1L5BY10 A0A267ESW3 A0A151WTD8 A0A2J7R478 A0A2J7QWK6 A0A2J7PW98 A0A2J7QP01 A0A2J7Q4S5 A0A0K2VK65 A0A151K0H9 A0A151K0I0 A0A151JXR4 A0A151JTI7 A0A151JZN7 A0A151JWC9 A0A151JWE8 A0A1W0WPR3 A0A2J7QND6 A0A151JUK9 A0A2J7RS37 A0A2J7R3C8 A0A151XI18 F4WPJ1 A0A151IRY2 A0A151JTQ3 A0A2J7PUI4 A0A2J7PJ78 A0A2J7R8T0 A0A151JU59 A0A195EWQ5 A0A151JAL8 B3CK30 A0A195EUP8 A0A195FM19 A0A2J7QJG3 A0A151WL87 A0A2J7PGR2 A0A2J7PRZ3 B3CK24 B3CK26 A0A026W2H3 B3CK19 A0A2J7QX91 A0A2J7QFV5 F4WVW5 A0A2J7PFM0 F4WY44 A0A2J7QVE3 A0A2J7Q656 A0A023F6W4 A0A2J7QQ11 A0A023F6P2 A0A2J7Q009 A0A2J7R0Z3 A0A2J7RQG1 A0A2J7PE56 F4X169 A0A2J7Q0F6 A0A2J7QR48
A0A023F6H6 A0A1L5BY10 A0A267ESW3 A0A151WTD8 A0A2J7R478 A0A2J7QWK6 A0A2J7PW98 A0A2J7QP01 A0A2J7Q4S5 A0A0K2VK65 A0A151K0H9 A0A151K0I0 A0A151JXR4 A0A151JTI7 A0A151JZN7 A0A151JWC9 A0A151JWE8 A0A1W0WPR3 A0A2J7QND6 A0A151JUK9 A0A2J7RS37 A0A2J7R3C8 A0A151XI18 F4WPJ1 A0A151IRY2 A0A151JTQ3 A0A2J7PUI4 A0A2J7PJ78 A0A2J7R8T0 A0A151JU59 A0A195EWQ5 A0A151JAL8 B3CK30 A0A195EUP8 A0A195FM19 A0A2J7QJG3 A0A151WL87 A0A2J7PGR2 A0A2J7PRZ3 B3CK24 B3CK26 A0A026W2H3 B3CK19 A0A2J7QX91 A0A2J7QFV5 F4WVW5 A0A2J7PFM0 F4WY44 A0A2J7QVE3 A0A2J7Q656 A0A023F6W4 A0A2J7QQ11 A0A023F6P2 A0A2J7Q009 A0A2J7R0Z3 A0A2J7RQG1 A0A2J7PE56 F4X169 A0A2J7Q0F6 A0A2J7QR48
PDB
3K9K
E-value=1.85283e-12,
Score=171
Ontologies
GO
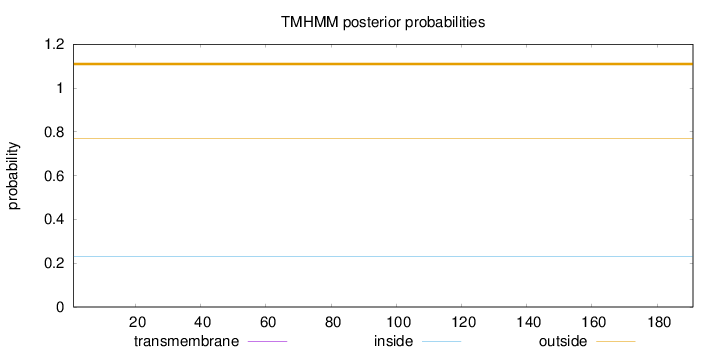
Topology
Length:
191
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.23018
outside
1 - 191
Population Genetic Test Statistics
Pi
28.060383
Theta
63.66158
Tajima's D
1.049555
CLR
65.071432
CSRT
0.67681615919204
Interpretation
Uncertain
