Gene
KWMTBOMO08906
Annotation
PREDICTED:_guanine_nucleotide-releasing_factor_2_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.253
Sequence
CDS
ATGGTGTTCGAGGATCGTCACGTCATTTCGGATCCTCCCGATCCACTAACGGTGCTTTTAGAGGGTGAAACCACTGGCGATCCAGGTTCAGGGCATCGTGGATCTCTGCGCGGTGCCAACAAGTTGGCGAGGCGAGCGCGATCCTTCAAAGACGATCTCCTCGAGCGTATTTCGAATATGCGTTCACCGGCTCAGCAGCACCATCAACCGCATCTTTCATCTGCTGTTCGTTCCCATTCACCCCTCAGTCCGAAGCTTCGAAATATGGGGGCGAAATCTAATACAGCCGAGGAGGGGGAAGTCACGCCCGCCAAAGAGTTGGAACGGCTCGTCAAAGAGGTAACATTCGGCTTGAAACACTTCTCGGACGTGATAACAAAGAGGAAATTGGAAATGCTTCCTGACAATGGCACGATCGTGTTCGAATCCATTGCGAACATACATAAAGCAATAAAACCGTACTGCAGTAGGTCGCCTCCTCTCACGAGCGCCGTTAAACGTCTTTGTGCGGCTCTCGCAAAGTTGATACGTCTGTGCGACGAAATCACCGCGGTCAGCTTACGCTCGGACGCCAGCAACGAGGAATTAGATACGTCGGTCGCAGCCCTGTCGCCCGATCACGTCAGTTCGATTGTTAAAGGCGTGACTTCCGCCGTGGAAGAAGTCGCCGCGTTAGCCCAACAGCACATGACGCGACCGGCGCTGTCGCCCCGACCGGCCCTCGTGTCGCCCAGATCCTCTACACAAAGAAATTCATTGCCAGATATACCATTAACACCCAAAGAACGTTCACTATTGACAGGTGAACCCGGCAATGAGGCAGGCGGCGTGCGCGCCTCCCATTCCTCGGAGTCGGTTCTGGACGCGCCCGACACGCCGCCCCCCAAACCTGCGAGACCCAATCGGAAAATCGAGTTCGCCCCACCTCTGCCGCCTAAGCGCAAGTCTAACAACGACAGCTCCCTACTGGGCTTGTCTATTGATAGGTTGTCTCTGCAGTCACACAGCAGCGGGTCCCTGGATTCTATGTTGAATATTTCGAACGACGAGGATAGGAACCACCATGACTCATCTAACCACGATAATGTTTTCATTTCGCAACAAAATAACGACGTTTCTGGGCTGTGTAGCTGCAGCAACGGAAATGACGTACGTACCTCAATGGAGTCACATGAATCACATCTGAACTCTGCTCACTTGAACTCGAACCATAATCGTTTCTCAAACGAGTCTGGATTCGTTTCTGTGGGACACGCTGAGATATCAACAGAGCACAGTTTCACGTCTTCATTCTCTTCGCATCATTACTCCAGCCATACGGATAGTACTTTTAACAGTACAGACTGTGTGAGCGAAATGAGGTGCACAGTGCAAAGTTTTGAGAGCCGCATGAATGTAACAAACATGAACCAATGTATCACGCACCATTCCTCGCAACTGGCGAGCATTACTTCAGACGAGAGCGAGACTCCACCGGAGCTGCCCGCCAAGACGAGGCGCAACATTCGAGCCGACGTACAGCAACACTTCCCGAATGTCGAAATCAGATCGAACCCAGTATGTTCTCTGCACGGCGATGCCAGACCGCACACATTAGCTGATCACCATCCACCGAGGCCCCCACCATTGCCGCTAAAGAAAAAACACATAATGGCGTACATGGAAATGTTCGGTAACTGTAGTCACCCGCCGAACAACGCGGCCGCACCGCCCGATATGTTCCGACATTCCATACACACGTACAATTTGGCCAGTTCTCAACACACAAGTGCAGGTGTGATTAGCGTTCAGAGACAACAAGTTCAAAGGTCTATAGCTCAATCGTTTACAGTACAACACTTTGGGAATCCGCCCTTGAGTGGGTCTGACGACAGTGTCAGCTTAACATCGAGCGGCACCAGTCCTTTACCTCTAGAACCGCCACCGGCTTTGCCGCCCAAGATGAATAAAAAACAGGTTCAAATGAATTCGAGCAACGAATTATTGCCTCCGCCGTCGCCGCGACCATCCTCCCACAATAGTTCGGCAGATGAAAGTCATTTAAATAATATTCAGTCCGATGAATCATTTATTAACATCACAACGCCTGGTAAATTCCCACTGGAATATGAGTTTGAGAATCTCTTGGAGACGTCGGCGCCGTCTCCTGCTCCGTCACAGAAAAGTGAAGAAGTTGTAGAGCCATCCCCTGTCGAAAAAGAAAAGCCCGCAACACCGACCCCACCTGACCCCGACGACATCGTGCTGCAGACCGACATCAACAAGTGGCTCGTACTGAAAGGTTCCAACAAGGATGGACCCGAAATACGCGGAGGTCACCCTGACGCTCTCATAGTGCTCGCGACTAAAGGAACCAAAGACTTCGCGTACCAGGAAGCGTTCCTGGCGACGTACCGCACGTGGGTGTCGCCGAGCGGGCTGGTGGCGCGGCTCGTGCGGCGCGCGCACCACCTGCGGGCGCGCCCCCACGACCTGCGCTCCACGCTCAGCCTGCTCACCAGGGTCGTCGCCGACCTCACGATGCTAGATTTGGATTACCCGTTAATGCAAGAGATAATGGAGTTCATCTACTGGTTGATTTACGAAGCCGAAGAGCTCGCAATAGCGAAAGCGCTGAGACAAATTCTAGTCGATAAACAGAAACAGTTGCATTTCCAACAAACAAACAATAGATACGAATACGACACGCCGTGCTACGGAACAGTGTCGCTTCGTCGCGATACGTTGTTGGATTTCAAAGCCACGGAAATAGCCGAACAGATGACGCTCTTGGACGCGGCGTTGTTTATCCGCATCACCACGGCCGAGGTGCTGTCGTGGCCTCGCGAGCAGTCCGAAGAGAGCTCACCCAATCTCACCCGATTCACGGAACACTTCAACAAGATGAGTTATTGGGCGCGGTCGAGGATCTTGGAACAGGAAAAGGCAATAGAACGTGAGAAATACGCCAGTAAATTCCTGAAAGTGATGAAGGCGTTACGCAAGATGAACAATTTCAATTCGTACCTGGCTCTGGTGTCCGCTTTGGACTCTCCGCCGGTGCGACGTCTCGGTTGGCCTCGATACATCGTCGACACTCTGCACGACTACTGCACCATCATAGACTCCTCGTCGTCTTTCCGCACCTACAGGCAAGCTTTAGCCGAGACGCAGCCGCCGTGTATACCGTACATCGGTCTCGTCTTGCAAGATTTAACTTTTGTCCATATTGGAAATCAAGATCTGTTATCGGATGGAAGTATTAATTTCACGAAGAGGTGGCAGCAGTATCACATCATGGAGAATATGAAGAGATTCCACAAAGACAGGCAGTACAAGTTCAAGAAGAACGAGCGCATCCAAGCCATGTTCAATGATTTCGACGACGTGCTGAGCGAGGAGGCGATGTGGCAGGTCTCCGAGAGCATCAAGCCGCGCGGCGGGAAGACTAAGCCGACCTCCGCCATTTGA
Protein
MVFEDRHVISDPPDPLTVLLEGETTGDPGSGHRGSLRGANKLARRARSFKDDLLERISNMRSPAQQHHQPHLSSAVRSHSPLSPKLRNMGAKSNTAEEGEVTPAKELERLVKEVTFGLKHFSDVITKRKLEMLPDNGTIVFESIANIHKAIKPYCSRSPPLTSAVKRLCAALAKLIRLCDEITAVSLRSDASNEELDTSVAALSPDHVSSIVKGVTSAVEEVAALAQQHMTRPALSPRPALVSPRSSTQRNSLPDIPLTPKERSLLTGEPGNEAGGVRASHSSESVLDAPDTPPPKPARPNRKIEFAPPLPPKRKSNNDSSLLGLSIDRLSLQSHSSGSLDSMLNISNDEDRNHHDSSNHDNVFISQQNNDVSGLCSCSNGNDVRTSMESHESHLNSAHLNSNHNRFSNESGFVSVGHAEISTEHSFTSSFSSHHYSSHTDSTFNSTDCVSEMRCTVQSFESRMNVTNMNQCITHHSSQLASITSDESETPPELPAKTRRNIRADVQQHFPNVEIRSNPVCSLHGDARPHTLADHHPPRPPPLPLKKKHIMAYMEMFGNCSHPPNNAAAPPDMFRHSIHTYNLASSQHTSAGVISVQRQQVQRSIAQSFTVQHFGNPPLSGSDDSVSLTSSGTSPLPLEPPPALPPKMNKKQVQMNSSNELLPPPSPRPSSHNSSADESHLNNIQSDESFINITTPGKFPLEYEFENLLETSAPSPAPSQKSEEVVEPSPVEKEKPATPTPPDPDDIVLQTDINKWLVLKGSNKDGPEIRGGHPDALIVLATKGTKDFAYQEAFLATYRTWVSPSGLVARLVRRAHHLRARPHDLRSTLSLLTRVVADLTMLDLDYPLMQEIMEFIYWLIYEAEELAIAKALRQILVDKQKQLHFQQTNNRYEYDTPCYGTVSLRRDTLLDFKATEIAEQMTLLDAALFIRITTAEVLSWPREQSEESSPNLTRFTEHFNKMSYWARSRILEQEKAIEREKYASKFLKVMKALRKMNNFNSYLALVSALDSPPVRRLGWPRYIVDTLHDYCTIIDSSSSFRTYRQALAETQPPCIPYIGLVLQDLTFVHIGNQDLLSDGSINFTKRWQQYHIMENMKRFHKDRQYKFKKNERIQAMFNDFDDVLSEEAMWQVSESIKPRGGKTKPTSAI
Summary
EMBL
Proteomes
Pfam
PF00617 RasGEF
Interpro
SUPFAM
SSF48366
SSF48366
Gene 3D
CDD
ProteinModelPortal
PDB
2IJE
E-value=5.74024e-25,
Score=288
Ontologies
GO
PANTHER
Topology
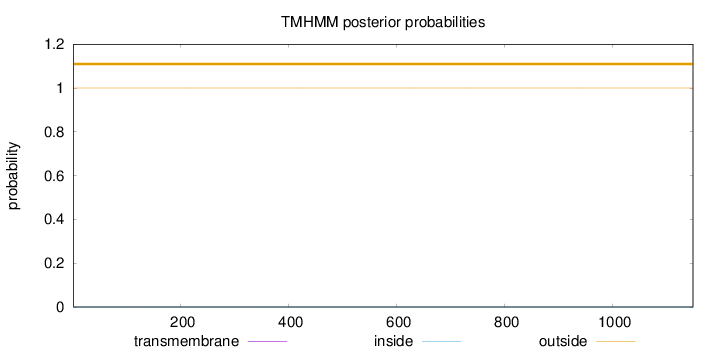
Length:
1149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00237
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1149
Population Genetic Test Statistics
Pi
270.428775
Theta
23.03021
Tajima's D
0.436416
CLR
0.63615
CSRT
0.498425078746063
Interpretation
Uncertain
