Pre Gene Modal
BGIBMGA007821
Annotation
hypothetical_protein_KGM_20642_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 2.825
Sequence
CDS
ATGGCGGACCCACTCAGTTTACTTCGTCAGTATAACGTAAATAAAAGAGAAATAATTGAACGCGATAACCAAATAATATTCGGTGAATTCTCTTGGCCAAAAAATGTTAAGACAAACTATCTTATTTGGAGATCTGGAAAGGAAGGCAGCGTTAAAGAGTATTACACCTTGGAATGCCTTCTTTTTATTCTCAAGAACATTCACCTGACTCATCCAGTATATGTCAGGCAGGCTGCTGCTGCAAATATACCACCTGTGCGTCGTCCAGATCGTAAAGAGCTATTAGCATATCTAAATGGAGAGACTGCTACTTGTGCCTCTATTGATAAAAGTGCACCCCTTGAAATTCCGACTCAAGTGAAACGCACACTTGATAATGATGGAGGAGAATCTGCTGCAAAAAAACCTCGAATTGAGGAAACTCATGTTCAGAAGGTCCGAGAACAACTGGCAGCCAGGCTTGATGCCCCTAAGGAAGCTTCAGTCACAGTGGACAATATCAAATCCCTTTCTGAAGCTATGTCTGTAGAAAAAATTGCAGCAATCAAAGCAAAACGATTAGCTAAGAAAAGAACCACAATCAAAAGCAATGACTATTCTGATACATTGGGTGTAGTCGGTTCAGACTTAAGAGCCATTCTTGACTATGATGTGGATCTTACTAAAGATATCATCAGCCGAGAAAGGCAGTGGCGGACAAGAACAACAGTATTACAAAGTAATGGGAAAATGTTTGCTAAAAGCATATTAGCTCATTTGGCGAGCATCAAAGCTAGAGAAGAAGGTAGACCTGGGATTCCGAGAGCACAACCTGTTATTTTGCAACAGCCTACTGTATTACCTGCACAACAGACACAGTATAACAGATACGATCAGGAAAGATTTATACGTCAAAAGGAAGAAACGGAAGGCTTCAAGATTGATACCATGGGTACCTATCATGGTATGACATTGAAATCAGTAACAGAAGGACCAAGTGTACCTGTAGCAGCTCGAGCCCCTCAAACGCCCAATCATCCAAGACAAATGCCACAAAACGGTGCTGTTATGCCTGGCACGCCTGGTCGCAGGGAGCTGCTGCCGGTGGCCGCAGCCCGACTTCCCGCAGGAGGAAAGCGGCCCTCACGCACTCCCATCATTATCATTCCCGCCGCAGCTACGTCACTCATCTCTATGTTTAATGTTAAAGATATTCTTCAAGATCTCAAATTTGTAACTGTAGAGCAAAAAAAAACTGAGGGAGCAGTTCGCGAGAATGAAGTACTTTTGCAAAGAAGAAAAGGTCAAACTGGCGATCAAATACCAACGAATGCCACTACAATAACGGTGCCATATCGAGTTGTAGACAATCCTGGAAGATTATCAGCAGCAGAATGGGACCGTGTTGTGGCGGTATTCGTGCAGGGACCTGCCTGGCAGTTTAAAGGATGGCCGTGGGACGGAAATCCAGTTCAAATCTTTGATAATATCTGTGCCTTCCACTTAAAGTATGATGAGATGAAGCTAGACGCGAACGTGGCGCGTTGGGCGGTCACTCTTCTCCAATTGAGTCGGACGAAGCGGCATTTGGATCGCGCCGTGTTACTCGGATTCTGGGAAACTCTCGACAAACATATGATGAAGAATAAACCACATCTTAGATTCTAA
Protein
MADPLSLLRQYNVNKREIIERDNQIIFGEFSWPKNVKTNYLIWRSGKEGSVKEYYTLECLLFILKNIHLTHPVYVRQAAAANIPPVRRPDRKELLAYLNGETATCASIDKSAPLEIPTQVKRTLDNDGGESAAKKPRIEETHVQKVREQLAARLDAPKEASVTVDNIKSLSEAMSVEKIAAIKAKRLAKKRTTIKSNDYSDTLGVVGSDLRAILDYDVDLTKDIISRERQWRTRTTVLQSNGKMFAKSILAHLASIKAREEGRPGIPRAQPVILQQPTVLPAQQTQYNRYDQERFIRQKEETEGFKIDTMGTYHGMTLKSVTEGPSVPVAARAPQTPNHPRQMPQNGAVMPGTPGRRELLPVAAARLPAGGKRPSRTPIIIIPAAATSLISMFNVKDILQDLKFVTVEQKKTEGAVRENEVLLQRRKGQTGDQIPTNATTITVPYRVVDNPGRLSAAEWDRVVAVFVQGPAWQFKGWPWDGNPVQIFDNICAFHLKYDEMKLDANVARWAVTLLQLSRTKRHLDRAVLLGFWETLDKHMMKNKPHLRF
Summary
Uniprot
H9JE74
A0A437AUM6
A0A2H1WVK5
A0A2A4K770
A0A212F405
A0A0N0PE50
+ More
A0A194PJQ3 A0A1W4X621 A0A2J7R1C4 D6X3J7 A0A1B6I6R7 A0A1B6EIT3 A0A0J7KTF2 E2ATY2 A0A1Y1MKG1 E9IKJ0 A0A154PRT7 A0A3L8DN32 F4WA25 E2C227 A0A2A3EFR2 A0A087ZYB3 A0A023F5E5 T1I333 A0A067R997 A0A0C9RFM0 A0A232EL14 K7J6P8 A0A0V0G7H9 A0A069DUI4 A0A2R7WW15 A0A026W0G6 A0A151IG05 A0A195DL20 A0A151I2H2 A0A151K007 A0A158P1B3 A0A151XHJ2 W8BT26 A0A182MKB1 A0A182RUP8 Q0IG22 A0A182Y930 A0A182P673 N6U2I4 E0VPB3 W5J929 A0A182Q9I5 A0A182VUA9 A0A2M4AGC8 A0A2M3ZHI9 A0A182F5S6 A0A0K8UPX8 A0A182NGM0 A0A2M4BJN9 A0A2M4BJG1 A0A2M4BJD5 A0A034WTR4 A0A182JSP6 A0A336MBK3 A0A1B0CUI6 A0A182XDC3 A0A182LMY3 Q5TNB0 A0A182I778 A0A0A1WKI3 A0A182TFH7 B4KB89 A0A182UPE1 A0A1Q3F3A9 B0WU18 A0A084VYN9 A0A182H4C3 A0A182JA25 A0A0L0CM84 B4M451 A0A1A9W9D4 A0A0P4VUT6 B4JFS1 A0A1A9VVX6 A0A1B0FMA2 A0A1A9XFK0 A0A1B0BXU2 A0A0A9WF39 B4PTZ6 A0A0N0BH28 B3P1U2 B3M1Z9 A0A1W4VQ87 A0A1L8DGP5 D0QWP3 A0A2H8TP93 Q294I1 B4GL28 A0A3B0JP66 Q9VHI1 B4QXA8 B4HKI5 A0A2S2Q414
A0A194PJQ3 A0A1W4X621 A0A2J7R1C4 D6X3J7 A0A1B6I6R7 A0A1B6EIT3 A0A0J7KTF2 E2ATY2 A0A1Y1MKG1 E9IKJ0 A0A154PRT7 A0A3L8DN32 F4WA25 E2C227 A0A2A3EFR2 A0A087ZYB3 A0A023F5E5 T1I333 A0A067R997 A0A0C9RFM0 A0A232EL14 K7J6P8 A0A0V0G7H9 A0A069DUI4 A0A2R7WW15 A0A026W0G6 A0A151IG05 A0A195DL20 A0A151I2H2 A0A151K007 A0A158P1B3 A0A151XHJ2 W8BT26 A0A182MKB1 A0A182RUP8 Q0IG22 A0A182Y930 A0A182P673 N6U2I4 E0VPB3 W5J929 A0A182Q9I5 A0A182VUA9 A0A2M4AGC8 A0A2M3ZHI9 A0A182F5S6 A0A0K8UPX8 A0A182NGM0 A0A2M4BJN9 A0A2M4BJG1 A0A2M4BJD5 A0A034WTR4 A0A182JSP6 A0A336MBK3 A0A1B0CUI6 A0A182XDC3 A0A182LMY3 Q5TNB0 A0A182I778 A0A0A1WKI3 A0A182TFH7 B4KB89 A0A182UPE1 A0A1Q3F3A9 B0WU18 A0A084VYN9 A0A182H4C3 A0A182JA25 A0A0L0CM84 B4M451 A0A1A9W9D4 A0A0P4VUT6 B4JFS1 A0A1A9VVX6 A0A1B0FMA2 A0A1A9XFK0 A0A1B0BXU2 A0A0A9WF39 B4PTZ6 A0A0N0BH28 B3P1U2 B3M1Z9 A0A1W4VQ87 A0A1L8DGP5 D0QWP3 A0A2H8TP93 Q294I1 B4GL28 A0A3B0JP66 Q9VHI1 B4QXA8 B4HKI5 A0A2S2Q414
Pubmed
19121390
22118469
26354079
18362917
19820115
20798317
+ More
28004739 21282665 30249741 21719571 25474469 24845553 28648823 20075255 26334808 24508170 21347285 24495485 17510324 25244985 23537049 20566863 20920257 23761445 25348373 20966253 12364791 14747013 17210077 25830018 17994087 18057021 24438588 26483478 26108605 27129103 25401762 26823975 17550304 19859648 15632085 10731132 12537568 12537572 12537573 12537574 16110336 16630820 17569856 17569867 26109357 26109356
28004739 21282665 30249741 21719571 25474469 24845553 28648823 20075255 26334808 24508170 21347285 24495485 17510324 25244985 23537049 20566863 20920257 23761445 25348373 20966253 12364791 14747013 17210077 25830018 17994087 18057021 24438588 26483478 26108605 27129103 25401762 26823975 17550304 19859648 15632085 10731132 12537568 12537572 12537573 12537574 16110336 16630820 17569856 17569867 26109357 26109356
EMBL
BABH01002701
RSAL01000401
RVE41917.1
ODYU01011338
SOQ57022.1
NWSH01000058
+ More
PCG80081.1 AGBW02010474 OWR48443.1 KQ459937 KPJ19215.1 KQ459602 KPI93258.1 NEVH01008208 PNF34640.1 KQ971374 EEZ97432.2 GECU01025082 JAS82624.1 GECZ01031932 JAS37837.1 LBMM01003399 KMQ93581.1 GL442747 EFN63113.1 GEZM01028884 JAV86213.1 GL763991 EFZ18901.1 KQ435090 KZC14583.1 QOIP01000006 RLU21238.1 GL888037 EGI68955.1 GL452067 EFN77978.1 KZ288266 PBC30122.1 GBBI01002353 JAC16359.1 ACPB03006536 KK852804 KDR16164.1 GBYB01005766 JAG75533.1 NNAY01003649 OXU19054.1 GECL01002065 JAP04059.1 GBGD01001106 JAC87783.1 KK855748 PTY23736.1 KK107503 EZA49557.1 KQ977743 KYN00165.1 KQ980762 KYN13580.1 KQ976535 KYM81464.1 KQ981301 KYN43048.1 ADTU01006200 ADTU01006201 ADTU01006202 KQ982138 KYQ59680.1 GAMC01004223 JAC02333.1 AXCM01000336 CH477274 EAT45191.1 APGK01042225 KB741002 KB632309 ENN75770.1 ERL92028.1 DS235363 EEB15219.1 ADMH02001840 ETN60952.1 AXCN02002086 GGFK01006519 MBW39840.1 GGFM01007265 MBW28016.1 GDHF01023668 JAI28646.1 GGFJ01004030 MBW53171.1 GGFJ01004031 MBW53172.1 GGFJ01004029 MBW53170.1 GAKP01001784 JAC57168.1 UFQT01000848 SSX27626.1 AJWK01029232 AAAB01008984 EAL38961.1 APCN01003368 GBXI01014965 JAC99326.1 CH933806 EDW15793.1 KRG01737.1 GFDL01013036 JAV22009.1 DS232099 EDS34734.1 ATLV01018382 KE525231 KFB43083.1 JXUM01001213 JXUM01001214 JXUM01001215 JXUM01030305 JXUM01030306 KQ560880 KXJ80501.1 JRES01000301 KNC32569.1 CH940652 EDW59412.1 GDKW01000266 JAI56329.1 CH916369 EDV93552.1 CCAG010013603 JXJN01022355 GBHO01038511 GBRD01015596 GDHC01004062 JAG05093.1 JAG50230.1 JAQ14567.1 CM000160 EDW96607.1 KQ435760 KOX75631.1 CH954181 EDV49830.1 CH902617 EDV43323.1 GFDF01008458 JAV05626.1 FJ821033 ACN94742.1 GFXV01004209 MBW16014.1 CM000070 EAL28983.1 CH479185 EDW38252.1 OUUW01000008 SPP84017.1 AE014297 AY061525 DQ305403 AAF54331.1 AAL29073.1 ABD58935.1 ACZ94861.1 CM000364 EDX13679.1 CH480815 EDW42935.1 GGMS01003292 MBY72495.1
PCG80081.1 AGBW02010474 OWR48443.1 KQ459937 KPJ19215.1 KQ459602 KPI93258.1 NEVH01008208 PNF34640.1 KQ971374 EEZ97432.2 GECU01025082 JAS82624.1 GECZ01031932 JAS37837.1 LBMM01003399 KMQ93581.1 GL442747 EFN63113.1 GEZM01028884 JAV86213.1 GL763991 EFZ18901.1 KQ435090 KZC14583.1 QOIP01000006 RLU21238.1 GL888037 EGI68955.1 GL452067 EFN77978.1 KZ288266 PBC30122.1 GBBI01002353 JAC16359.1 ACPB03006536 KK852804 KDR16164.1 GBYB01005766 JAG75533.1 NNAY01003649 OXU19054.1 GECL01002065 JAP04059.1 GBGD01001106 JAC87783.1 KK855748 PTY23736.1 KK107503 EZA49557.1 KQ977743 KYN00165.1 KQ980762 KYN13580.1 KQ976535 KYM81464.1 KQ981301 KYN43048.1 ADTU01006200 ADTU01006201 ADTU01006202 KQ982138 KYQ59680.1 GAMC01004223 JAC02333.1 AXCM01000336 CH477274 EAT45191.1 APGK01042225 KB741002 KB632309 ENN75770.1 ERL92028.1 DS235363 EEB15219.1 ADMH02001840 ETN60952.1 AXCN02002086 GGFK01006519 MBW39840.1 GGFM01007265 MBW28016.1 GDHF01023668 JAI28646.1 GGFJ01004030 MBW53171.1 GGFJ01004031 MBW53172.1 GGFJ01004029 MBW53170.1 GAKP01001784 JAC57168.1 UFQT01000848 SSX27626.1 AJWK01029232 AAAB01008984 EAL38961.1 APCN01003368 GBXI01014965 JAC99326.1 CH933806 EDW15793.1 KRG01737.1 GFDL01013036 JAV22009.1 DS232099 EDS34734.1 ATLV01018382 KE525231 KFB43083.1 JXUM01001213 JXUM01001214 JXUM01001215 JXUM01030305 JXUM01030306 KQ560880 KXJ80501.1 JRES01000301 KNC32569.1 CH940652 EDW59412.1 GDKW01000266 JAI56329.1 CH916369 EDV93552.1 CCAG010013603 JXJN01022355 GBHO01038511 GBRD01015596 GDHC01004062 JAG05093.1 JAG50230.1 JAQ14567.1 CM000160 EDW96607.1 KQ435760 KOX75631.1 CH954181 EDV49830.1 CH902617 EDV43323.1 GFDF01008458 JAV05626.1 FJ821033 ACN94742.1 GFXV01004209 MBW16014.1 CM000070 EAL28983.1 CH479185 EDW38252.1 OUUW01000008 SPP84017.1 AE014297 AY061525 DQ305403 AAF54331.1 AAL29073.1 ABD58935.1 ACZ94861.1 CM000364 EDX13679.1 CH480815 EDW42935.1 GGMS01003292 MBY72495.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000053268
+ More
UP000192223 UP000235965 UP000007266 UP000036403 UP000000311 UP000076502 UP000279307 UP000007755 UP000008237 UP000242457 UP000005203 UP000015103 UP000027135 UP000215335 UP000002358 UP000053097 UP000078542 UP000078492 UP000078540 UP000078541 UP000005205 UP000075809 UP000075883 UP000075900 UP000008820 UP000076408 UP000075885 UP000019118 UP000030742 UP000009046 UP000000673 UP000075886 UP000075920 UP000069272 UP000075884 UP000075881 UP000092461 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000009192 UP000075903 UP000002320 UP000030765 UP000069940 UP000249989 UP000075880 UP000037069 UP000008792 UP000091820 UP000001070 UP000078200 UP000092444 UP000092443 UP000092460 UP000002282 UP000053105 UP000008711 UP000007801 UP000192221 UP000001819 UP000008744 UP000268350 UP000000803 UP000000304 UP000001292
UP000192223 UP000235965 UP000007266 UP000036403 UP000000311 UP000076502 UP000279307 UP000007755 UP000008237 UP000242457 UP000005203 UP000015103 UP000027135 UP000215335 UP000002358 UP000053097 UP000078542 UP000078492 UP000078540 UP000078541 UP000005205 UP000075809 UP000075883 UP000075900 UP000008820 UP000076408 UP000075885 UP000019118 UP000030742 UP000009046 UP000000673 UP000075886 UP000075920 UP000069272 UP000075884 UP000075881 UP000092461 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000009192 UP000075903 UP000002320 UP000030765 UP000069940 UP000249989 UP000075880 UP000037069 UP000008792 UP000091820 UP000001070 UP000078200 UP000092444 UP000092443 UP000092460 UP000002282 UP000053105 UP000008711 UP000007801 UP000192221 UP000001819 UP000008744 UP000268350 UP000000803 UP000000304 UP000001292
Interpro
Gene 3D
ProteinModelPortal
H9JE74
A0A437AUM6
A0A2H1WVK5
A0A2A4K770
A0A212F405
A0A0N0PE50
+ More
A0A194PJQ3 A0A1W4X621 A0A2J7R1C4 D6X3J7 A0A1B6I6R7 A0A1B6EIT3 A0A0J7KTF2 E2ATY2 A0A1Y1MKG1 E9IKJ0 A0A154PRT7 A0A3L8DN32 F4WA25 E2C227 A0A2A3EFR2 A0A087ZYB3 A0A023F5E5 T1I333 A0A067R997 A0A0C9RFM0 A0A232EL14 K7J6P8 A0A0V0G7H9 A0A069DUI4 A0A2R7WW15 A0A026W0G6 A0A151IG05 A0A195DL20 A0A151I2H2 A0A151K007 A0A158P1B3 A0A151XHJ2 W8BT26 A0A182MKB1 A0A182RUP8 Q0IG22 A0A182Y930 A0A182P673 N6U2I4 E0VPB3 W5J929 A0A182Q9I5 A0A182VUA9 A0A2M4AGC8 A0A2M3ZHI9 A0A182F5S6 A0A0K8UPX8 A0A182NGM0 A0A2M4BJN9 A0A2M4BJG1 A0A2M4BJD5 A0A034WTR4 A0A182JSP6 A0A336MBK3 A0A1B0CUI6 A0A182XDC3 A0A182LMY3 Q5TNB0 A0A182I778 A0A0A1WKI3 A0A182TFH7 B4KB89 A0A182UPE1 A0A1Q3F3A9 B0WU18 A0A084VYN9 A0A182H4C3 A0A182JA25 A0A0L0CM84 B4M451 A0A1A9W9D4 A0A0P4VUT6 B4JFS1 A0A1A9VVX6 A0A1B0FMA2 A0A1A9XFK0 A0A1B0BXU2 A0A0A9WF39 B4PTZ6 A0A0N0BH28 B3P1U2 B3M1Z9 A0A1W4VQ87 A0A1L8DGP5 D0QWP3 A0A2H8TP93 Q294I1 B4GL28 A0A3B0JP66 Q9VHI1 B4QXA8 B4HKI5 A0A2S2Q414
A0A194PJQ3 A0A1W4X621 A0A2J7R1C4 D6X3J7 A0A1B6I6R7 A0A1B6EIT3 A0A0J7KTF2 E2ATY2 A0A1Y1MKG1 E9IKJ0 A0A154PRT7 A0A3L8DN32 F4WA25 E2C227 A0A2A3EFR2 A0A087ZYB3 A0A023F5E5 T1I333 A0A067R997 A0A0C9RFM0 A0A232EL14 K7J6P8 A0A0V0G7H9 A0A069DUI4 A0A2R7WW15 A0A026W0G6 A0A151IG05 A0A195DL20 A0A151I2H2 A0A151K007 A0A158P1B3 A0A151XHJ2 W8BT26 A0A182MKB1 A0A182RUP8 Q0IG22 A0A182Y930 A0A182P673 N6U2I4 E0VPB3 W5J929 A0A182Q9I5 A0A182VUA9 A0A2M4AGC8 A0A2M3ZHI9 A0A182F5S6 A0A0K8UPX8 A0A182NGM0 A0A2M4BJN9 A0A2M4BJG1 A0A2M4BJD5 A0A034WTR4 A0A182JSP6 A0A336MBK3 A0A1B0CUI6 A0A182XDC3 A0A182LMY3 Q5TNB0 A0A182I778 A0A0A1WKI3 A0A182TFH7 B4KB89 A0A182UPE1 A0A1Q3F3A9 B0WU18 A0A084VYN9 A0A182H4C3 A0A182JA25 A0A0L0CM84 B4M451 A0A1A9W9D4 A0A0P4VUT6 B4JFS1 A0A1A9VVX6 A0A1B0FMA2 A0A1A9XFK0 A0A1B0BXU2 A0A0A9WF39 B4PTZ6 A0A0N0BH28 B3P1U2 B3M1Z9 A0A1W4VQ87 A0A1L8DGP5 D0QWP3 A0A2H8TP93 Q294I1 B4GL28 A0A3B0JP66 Q9VHI1 B4QXA8 B4HKI5 A0A2S2Q414
PDB
5YDE
E-value=3.1328e-42,
Score=434
Ontologies
GO
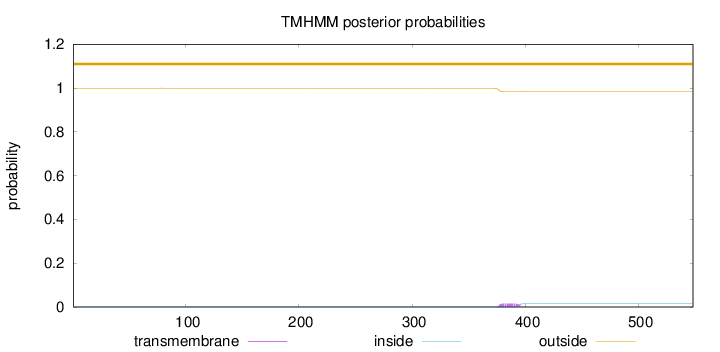
Topology
Length:
548
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28931
Exp number, first 60 AAs:
0.00048
Total prob of N-in:
0.00067
outside
1 - 548
Population Genetic Test Statistics
Pi
232.213923
Theta
165.756773
Tajima's D
0.508766
CLR
0.093516
CSRT
0.515574221288936
Interpretation
Uncertain
