Gene
KWMTBOMO08904
Pre Gene Modal
BGIBMGA007713
Annotation
hypothetical_protein_KGM_20648_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 4.972
Sequence
CDS
ATGCGTGATAAAATCGCAGCATTGTTTCGAGGCTCGGCGCACACACCTGCACACAACACAGAAAAGCTTCAGAATGGTGTGCTTGATCCAGTACTCGTCTTTTCGGAAGGGAGCACACGCGAAGGCGGATTGGTTTGGCGCGGGTGCTCAGCCAGCTGCGATGCCGAAGACGGCACGACGGGAGCGTTCGACGATGGCAACTCAAACCCAGGAGACAAGCTGGAGGGGTCGGACGCAGCTCCGGACGACCCTGTTCACCTCAAGAGACGGGTGGGACTCTTCAGTGGGGTGGCTCTAATTGTCGGCACTATGATTGGATCAGGAATATTCGTATCGCCCTCTGGGCTTTTGGCGCGTACCGGCTCAGTGGGCATAAGCTTCATCATTTGGATGGCATGTGGCCTGCTCTCGTTGCTTGGAGCGCTTGCGTATGCGGAATTAGGGACAATGAATACATCATCTGGCGCTGAATACGCCTATTTCATGGACGCCTTCGGGGGCCCACCCGCATTCCTCTTTTCATGGGTATCGACATTGGTTTTAAAACCATCACAAATGGCCATTATTTGCCTAAGCTTCGCCAAATACGCCGTTGAGCCTTTTGTAGCGGAGTGCGAACCCCCTGACAGTTTGGTCAAATTGGTTGCGGTCATTTCGATTGTGATGATCTTAGCAGTGAATTGCTACAGTGTCAACTTGGCAACGAATGTGCAAAATATTTTCACAGCGGCCAAACTCGTTGCGATCGCGATCATCGTATGCGGAGGAGCTTACAAACTGATACTAGGTAATACGCGACATTTACAGGAGCCCAACTTCGCCAGTAGCAAGGCGACTCTGGGCAACATTGCCACCGCATTCTACACCGGCCTATGGGCTTACGACGGATGGAATAACCTTAATTATGTCACGGAAGAAATAAAAAATCCTTCCAAGAATCTCCCGCTAAGTATCATAATCGGGATACCGTTAGTAACACTATGCTACGCTCTTGTGAATGTATCATATTTGGCGGTGATGTCCGTGAGCGAGATGGCGGATAGTGAGGCAGTGGCGGTCACATTCGGTAACCGATTGCTCGGCCCGATGGCCTGGTTGATGCCATTAGCCGTCACTATATCTACTTTTGGATCAGCGAATGGAACTCTATTCGTGGCCGGAAGGTTGTGTTTTGCCGCGTCACGAGAAGGTCACTTATTGGACATCCTCTCTTACGTGCACGTGCGCCGTTTCACACCAGCACCCGGATTGATATTCCACTCTTTAATAGCCGTGGCAATGGTGTTGTACGGGACGATAGATTCGTTAATAGATTTCTTCTCGTTCACCGCTTGGATATTCTACGGAGGCGCGATGCTCGCTTTGATTGTGATGCGGTATACGAAACCGCACGCCCCAAGACCTTACAAGGTACCAATAATTATTCCGTACATCGTGCTGTTTGTATCGGCGTATCTCGTGGTGGCTCCTATACTGGACAAGCCTCAGTGGGAGTACATGTATGCGGCAGCGTTTATATTGGGAGGGCTGCTCGTGTATCTTCCGTTCGTCAAATGGGGATATTCATTGCCGTTTATGGATAAAATTACGGTGTTTCTGCAAATGGTACTAGAGGTAGTGCCAACATCTACAACGTTCGAATACTAA
Protein
MRDKIAALFRGSAHTPAHNTEKLQNGVLDPVLVFSEGSTREGGLVWRGCSASCDAEDGTTGAFDDGNSNPGDKLEGSDAAPDDPVHLKRRVGLFSGVALIVGTMIGSGIFVSPSGLLARTGSVGISFIIWMACGLLSLLGALAYAELGTMNTSSGAEYAYFMDAFGGPPAFLFSWVSTLVLKPSQMAIICLSFAKYAVEPFVAECEPPDSLVKLVAVISIVMILAVNCYSVNLATNVQNIFTAAKLVAIAIIVCGGAYKLILGNTRHLQEPNFASSKATLGNIATAFYTGLWAYDGWNNLNYVTEEIKNPSKNLPLSIIIGIPLVTLCYALVNVSYLAVMSVSEMADSEAVAVTFGNRLLGPMAWLMPLAVTISTFGSANGTLFVAGRLCFAASREGHLLDILSYVHVRRFTPAPGLIFHSLIAVAMVLYGTIDSLIDFFSFTAWIFYGGAMLALIVMRYTKPHAPRPYKVPIIIPYIVLFVSAYLVVAPILDKPQWEYMYAAAFILGGLLVYLPFVKWGYSLPFMDKITVFLQMVLEVVPTSTTFEY
Summary
Uniprot
A0A2H1WVD7
A0A212F3Y6
A0A437AUV0
A0A194PIW0
S4NIG5
A0A2A4K8U4
+ More
A0A0N1II68 A0A2J7QV32 D6W9G6 A0A139WMX9 A0A1Y1KDG1 A0A1Y1KDC5 A0A034VVL9 A0A1B6BWJ6 A0A1B6C3H6 A0A0A1WWN7 A0A1L8DEN8 A0A067QUR4 A0A182J0J2 W8BZ65 A0A182KM63 A0A182XBN6 Q17FJ5 A0A182Q991 A0A182VJP8 A0A1S4GDC1 A0A182I0H3 A0A182PEE0 A0A182N4W8 A0A1A9X029 A0A084VRU1 A0A034VX81 A0A1Q3G4V0 U5EVD8 Q7PUF8 N6T9S3 A0A0L0C5A0 A0A1A9UUZ9 A0A1A9ZLP1 W8C7J8 A0A1I8P8H0 V5GZR6 A0A1I8MTX2 A0A0V0GBA4 A0A224XII0 A0A023F445 A0A1B6FQJ0 A0A1Y1KGM3 A0A0P4VS77 A0A1Y1KH01 W8BPF5 A0A1I8P8N6 A0A1I8MTX1 A0A1B6FMC4 A0A1B6DUD2 A0A1Y1KIS8 A0A1B6CA36 A0A1B6D1N1 A0A1Y1KJE2 A0A1B6CNE9 A0A1B6DMR6 B4M2U8 A0A182MSY9 Q29H89 A0A182YJW2 A0A0Q9XGC2 A0A0R3P000 B4GY51 A0A1B6L6N5 A0A0Q9X4E6 A0A026W1X3 A0A3B0KJH7 A0A1B6KW92 A0A0Q9WBD2 A0A0M3QZQ7 E2BN13 K7J3G3 B4JL41 B4NPQ8 A0A0L7R509 A0A1B6GB68 A0A158NG72 A0A158NG73 A0A088A5G4 A0A1B6G1V2 A0A1J1HP88 A0A1B6FM64 A0A1J1HN14 A0A1B6FJV7 A0A1W4VVJ0 E2A8N6 C9QPC3 A0A310SJX9 B4IGE5 A0A0R1EAL2 A0A0Q5TFN2 Q8IR48 A0A0A9Z174 A0A146L4D7 A0A1B0CBF0
A0A0N1II68 A0A2J7QV32 D6W9G6 A0A139WMX9 A0A1Y1KDG1 A0A1Y1KDC5 A0A034VVL9 A0A1B6BWJ6 A0A1B6C3H6 A0A0A1WWN7 A0A1L8DEN8 A0A067QUR4 A0A182J0J2 W8BZ65 A0A182KM63 A0A182XBN6 Q17FJ5 A0A182Q991 A0A182VJP8 A0A1S4GDC1 A0A182I0H3 A0A182PEE0 A0A182N4W8 A0A1A9X029 A0A084VRU1 A0A034VX81 A0A1Q3G4V0 U5EVD8 Q7PUF8 N6T9S3 A0A0L0C5A0 A0A1A9UUZ9 A0A1A9ZLP1 W8C7J8 A0A1I8P8H0 V5GZR6 A0A1I8MTX2 A0A0V0GBA4 A0A224XII0 A0A023F445 A0A1B6FQJ0 A0A1Y1KGM3 A0A0P4VS77 A0A1Y1KH01 W8BPF5 A0A1I8P8N6 A0A1I8MTX1 A0A1B6FMC4 A0A1B6DUD2 A0A1Y1KIS8 A0A1B6CA36 A0A1B6D1N1 A0A1Y1KJE2 A0A1B6CNE9 A0A1B6DMR6 B4M2U8 A0A182MSY9 Q29H89 A0A182YJW2 A0A0Q9XGC2 A0A0R3P000 B4GY51 A0A1B6L6N5 A0A0Q9X4E6 A0A026W1X3 A0A3B0KJH7 A0A1B6KW92 A0A0Q9WBD2 A0A0M3QZQ7 E2BN13 K7J3G3 B4JL41 B4NPQ8 A0A0L7R509 A0A1B6GB68 A0A158NG72 A0A158NG73 A0A088A5G4 A0A1B6G1V2 A0A1J1HP88 A0A1B6FM64 A0A1J1HN14 A0A1B6FJV7 A0A1W4VVJ0 E2A8N6 C9QPC3 A0A310SJX9 B4IGE5 A0A0R1EAL2 A0A0Q5TFN2 Q8IR48 A0A0A9Z174 A0A146L4D7 A0A1B0CBF0
Pubmed
22118469
26354079
23622113
18362917
19820115
28004739
+ More
25348373 25830018 24845553 24495485 20966253 17510324 12364791 24438588 23537049 26108605 25315136 25474469 27129103 17994087 15632085 25244985 24508170 20798317 20075255 21347285 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25401762 26823975
25348373 25830018 24845553 24495485 20966253 17510324 12364791 24438588 23537049 26108605 25315136 25474469 27129103 17994087 15632085 25244985 24508170 20798317 20075255 21347285 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25401762 26823975
EMBL
ODYU01011338
SOQ57021.1
AGBW02010474
OWR48444.1
RSAL01000401
RVE41916.1
+ More
KQ459602 KPI93257.1 GAIX01014084 JAA78476.1 NWSH01000058 PCG80082.1 KQ459937 KPJ19214.1 NEVH01010479 PNF32443.1 KQ971312 EEZ98157.2 KYB29191.1 GEZM01086362 JAV59519.1 GEZM01086365 JAV59513.1 GAKP01012815 JAC46137.1 GEDC01031642 JAS05656.1 GEDC01029251 JAS08047.1 GBXI01010813 JAD03479.1 GFDF01009173 JAV04911.1 KK852927 KDR13693.1 GAMC01007969 GAMC01007968 JAB98586.1 CH477270 EAT45348.1 AXCN02000187 AAAB01008987 APCN01002077 APCN01002078 ATLV01015780 KE525036 KFB40685.1 GAKP01012814 JAC46138.1 GFDL01000221 JAV34824.1 GANO01001904 JAB57967.1 EAA01594.6 APGK01046275 KB741050 KB631815 ENN74488.1 ERL86481.1 JRES01000902 KNC27406.1 GAMC01007966 JAB98589.1 GALX01002443 JAB66023.1 GECL01001543 JAP04581.1 GFTR01006798 JAW09628.1 GBBI01002447 JAC16265.1 GECZ01017292 JAS52477.1 GEZM01086361 JAV59521.1 GDKW01001817 JAI54778.1 GEZM01086366 JAV59510.1 GAMC01007967 JAB98588.1 GECZ01018439 JAS51330.1 GEDC01008008 JAS29290.1 GEZM01086364 JAV59515.1 GEDC01026950 JAS10348.1 GEDC01017687 JAS19611.1 GEZM01086363 JAV59516.1 GEDC01022354 JAS14944.1 GEDC01010340 JAS26958.1 CH940651 EDW65123.1 AXCM01005021 AXCM01005022 CH379064 EAL31869.1 CH933813 KRG07389.1 KRT06619.1 CH479196 EDW27678.1 GEBQ01020605 JAT19372.1 CH964291 KRG00388.1 KK107487 EZA50033.1 OUUW01000015 SPP88710.1 GEBQ01024316 JAT15661.1 KRF82028.1 CP012528 ALC49799.1 GL449382 EFN82917.1 CH916370 EDW00294.1 EDW86498.1 KQ414654 KOC65967.1 GECZ01010107 JAS59662.1 ADTU01015040 GECZ01013359 JAS56410.1 CVRI01000012 CRK89346.1 GECZ01018492 JAS51277.1 CRK89347.1 GECZ01019304 JAS50465.1 GL437652 EFN70223.1 BT100005 ACX54889.1 KQ761783 OAD56986.1 CH480835 EDW48879.1 CM000162 KRK06297.1 CH954180 KQS30341.1 AE014298 AAN09339.1 GBHO01004577 JAG39027.1 GDHC01016989 JAQ01640.1 AJWK01005094 AJWK01005095
KQ459602 KPI93257.1 GAIX01014084 JAA78476.1 NWSH01000058 PCG80082.1 KQ459937 KPJ19214.1 NEVH01010479 PNF32443.1 KQ971312 EEZ98157.2 KYB29191.1 GEZM01086362 JAV59519.1 GEZM01086365 JAV59513.1 GAKP01012815 JAC46137.1 GEDC01031642 JAS05656.1 GEDC01029251 JAS08047.1 GBXI01010813 JAD03479.1 GFDF01009173 JAV04911.1 KK852927 KDR13693.1 GAMC01007969 GAMC01007968 JAB98586.1 CH477270 EAT45348.1 AXCN02000187 AAAB01008987 APCN01002077 APCN01002078 ATLV01015780 KE525036 KFB40685.1 GAKP01012814 JAC46138.1 GFDL01000221 JAV34824.1 GANO01001904 JAB57967.1 EAA01594.6 APGK01046275 KB741050 KB631815 ENN74488.1 ERL86481.1 JRES01000902 KNC27406.1 GAMC01007966 JAB98589.1 GALX01002443 JAB66023.1 GECL01001543 JAP04581.1 GFTR01006798 JAW09628.1 GBBI01002447 JAC16265.1 GECZ01017292 JAS52477.1 GEZM01086361 JAV59521.1 GDKW01001817 JAI54778.1 GEZM01086366 JAV59510.1 GAMC01007967 JAB98588.1 GECZ01018439 JAS51330.1 GEDC01008008 JAS29290.1 GEZM01086364 JAV59515.1 GEDC01026950 JAS10348.1 GEDC01017687 JAS19611.1 GEZM01086363 JAV59516.1 GEDC01022354 JAS14944.1 GEDC01010340 JAS26958.1 CH940651 EDW65123.1 AXCM01005021 AXCM01005022 CH379064 EAL31869.1 CH933813 KRG07389.1 KRT06619.1 CH479196 EDW27678.1 GEBQ01020605 JAT19372.1 CH964291 KRG00388.1 KK107487 EZA50033.1 OUUW01000015 SPP88710.1 GEBQ01024316 JAT15661.1 KRF82028.1 CP012528 ALC49799.1 GL449382 EFN82917.1 CH916370 EDW00294.1 EDW86498.1 KQ414654 KOC65967.1 GECZ01010107 JAS59662.1 ADTU01015040 GECZ01013359 JAS56410.1 CVRI01000012 CRK89346.1 GECZ01018492 JAS51277.1 CRK89347.1 GECZ01019304 JAS50465.1 GL437652 EFN70223.1 BT100005 ACX54889.1 KQ761783 OAD56986.1 CH480835 EDW48879.1 CM000162 KRK06297.1 CH954180 KQS30341.1 AE014298 AAN09339.1 GBHO01004577 JAG39027.1 GDHC01016989 JAQ01640.1 AJWK01005094 AJWK01005095
Proteomes
UP000007151
UP000283053
UP000053268
UP000218220
UP000053240
UP000235965
+ More
UP000007266 UP000027135 UP000075880 UP000075882 UP000076407 UP000008820 UP000075886 UP000075903 UP000075840 UP000075885 UP000075884 UP000091820 UP000030765 UP000007062 UP000019118 UP000030742 UP000037069 UP000078200 UP000092445 UP000095300 UP000095301 UP000008792 UP000075883 UP000001819 UP000076408 UP000009192 UP000008744 UP000007798 UP000053097 UP000268350 UP000092553 UP000008237 UP000002358 UP000001070 UP000053825 UP000005205 UP000005203 UP000183832 UP000192221 UP000000311 UP000001292 UP000002282 UP000008711 UP000000803 UP000092461
UP000007266 UP000027135 UP000075880 UP000075882 UP000076407 UP000008820 UP000075886 UP000075903 UP000075840 UP000075885 UP000075884 UP000091820 UP000030765 UP000007062 UP000019118 UP000030742 UP000037069 UP000078200 UP000092445 UP000095300 UP000095301 UP000008792 UP000075883 UP000001819 UP000076408 UP000009192 UP000008744 UP000007798 UP000053097 UP000268350 UP000092553 UP000008237 UP000002358 UP000001070 UP000053825 UP000005205 UP000005203 UP000183832 UP000192221 UP000000311 UP000001292 UP000002282 UP000008711 UP000000803 UP000092461
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
A0A2H1WVD7
A0A212F3Y6
A0A437AUV0
A0A194PIW0
S4NIG5
A0A2A4K8U4
+ More
A0A0N1II68 A0A2J7QV32 D6W9G6 A0A139WMX9 A0A1Y1KDG1 A0A1Y1KDC5 A0A034VVL9 A0A1B6BWJ6 A0A1B6C3H6 A0A0A1WWN7 A0A1L8DEN8 A0A067QUR4 A0A182J0J2 W8BZ65 A0A182KM63 A0A182XBN6 Q17FJ5 A0A182Q991 A0A182VJP8 A0A1S4GDC1 A0A182I0H3 A0A182PEE0 A0A182N4W8 A0A1A9X029 A0A084VRU1 A0A034VX81 A0A1Q3G4V0 U5EVD8 Q7PUF8 N6T9S3 A0A0L0C5A0 A0A1A9UUZ9 A0A1A9ZLP1 W8C7J8 A0A1I8P8H0 V5GZR6 A0A1I8MTX2 A0A0V0GBA4 A0A224XII0 A0A023F445 A0A1B6FQJ0 A0A1Y1KGM3 A0A0P4VS77 A0A1Y1KH01 W8BPF5 A0A1I8P8N6 A0A1I8MTX1 A0A1B6FMC4 A0A1B6DUD2 A0A1Y1KIS8 A0A1B6CA36 A0A1B6D1N1 A0A1Y1KJE2 A0A1B6CNE9 A0A1B6DMR6 B4M2U8 A0A182MSY9 Q29H89 A0A182YJW2 A0A0Q9XGC2 A0A0R3P000 B4GY51 A0A1B6L6N5 A0A0Q9X4E6 A0A026W1X3 A0A3B0KJH7 A0A1B6KW92 A0A0Q9WBD2 A0A0M3QZQ7 E2BN13 K7J3G3 B4JL41 B4NPQ8 A0A0L7R509 A0A1B6GB68 A0A158NG72 A0A158NG73 A0A088A5G4 A0A1B6G1V2 A0A1J1HP88 A0A1B6FM64 A0A1J1HN14 A0A1B6FJV7 A0A1W4VVJ0 E2A8N6 C9QPC3 A0A310SJX9 B4IGE5 A0A0R1EAL2 A0A0Q5TFN2 Q8IR48 A0A0A9Z174 A0A146L4D7 A0A1B0CBF0
A0A0N1II68 A0A2J7QV32 D6W9G6 A0A139WMX9 A0A1Y1KDG1 A0A1Y1KDC5 A0A034VVL9 A0A1B6BWJ6 A0A1B6C3H6 A0A0A1WWN7 A0A1L8DEN8 A0A067QUR4 A0A182J0J2 W8BZ65 A0A182KM63 A0A182XBN6 Q17FJ5 A0A182Q991 A0A182VJP8 A0A1S4GDC1 A0A182I0H3 A0A182PEE0 A0A182N4W8 A0A1A9X029 A0A084VRU1 A0A034VX81 A0A1Q3G4V0 U5EVD8 Q7PUF8 N6T9S3 A0A0L0C5A0 A0A1A9UUZ9 A0A1A9ZLP1 W8C7J8 A0A1I8P8H0 V5GZR6 A0A1I8MTX2 A0A0V0GBA4 A0A224XII0 A0A023F445 A0A1B6FQJ0 A0A1Y1KGM3 A0A0P4VS77 A0A1Y1KH01 W8BPF5 A0A1I8P8N6 A0A1I8MTX1 A0A1B6FMC4 A0A1B6DUD2 A0A1Y1KIS8 A0A1B6CA36 A0A1B6D1N1 A0A1Y1KJE2 A0A1B6CNE9 A0A1B6DMR6 B4M2U8 A0A182MSY9 Q29H89 A0A182YJW2 A0A0Q9XGC2 A0A0R3P000 B4GY51 A0A1B6L6N5 A0A0Q9X4E6 A0A026W1X3 A0A3B0KJH7 A0A1B6KW92 A0A0Q9WBD2 A0A0M3QZQ7 E2BN13 K7J3G3 B4JL41 B4NPQ8 A0A0L7R509 A0A1B6GB68 A0A158NG72 A0A158NG73 A0A088A5G4 A0A1B6G1V2 A0A1J1HP88 A0A1B6FM64 A0A1J1HN14 A0A1B6FJV7 A0A1W4VVJ0 E2A8N6 C9QPC3 A0A310SJX9 B4IGE5 A0A0R1EAL2 A0A0Q5TFN2 Q8IR48 A0A0A9Z174 A0A146L4D7 A0A1B0CBF0
PDB
6IRT
E-value=9.62352e-85,
Score=800
Ontologies
GO
Topology
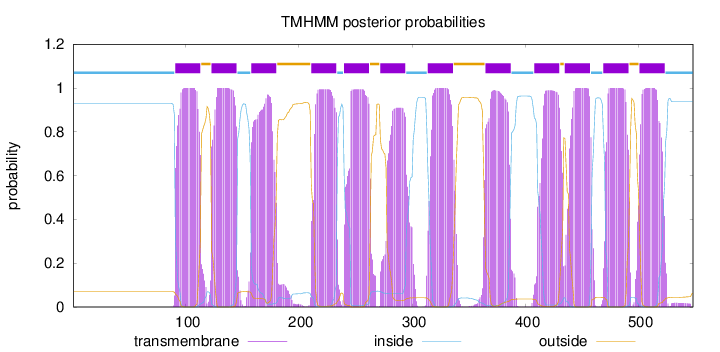
Length:
548
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
258.60975
Exp number, first 60 AAs:
0
Total prob of N-in:
0.92990
inside
1 - 90
TMhelix
91 - 113
outside
114 - 122
TMhelix
123 - 145
inside
146 - 157
TMhelix
158 - 180
outside
181 - 210
TMhelix
211 - 233
inside
234 - 239
TMhelix
240 - 262
outside
263 - 271
TMhelix
272 - 294
inside
295 - 313
TMhelix
314 - 336
outside
337 - 364
TMhelix
365 - 387
inside
388 - 407
TMhelix
408 - 430
outside
431 - 434
TMhelix
435 - 457
inside
458 - 468
TMhelix
469 - 491
outside
492 - 500
TMhelix
501 - 523
inside
524 - 548
Population Genetic Test Statistics
Pi
210.089107
Theta
176.401547
Tajima's D
0.631805
CLR
0.103121
CSRT
0.55697215139243
Interpretation
Uncertain
